Abstract
Deficiencies in the IL-7 signaling pathway result in severe disruptions of lymphoid development in adult mice. To understand more about how IL-7 deficiency impacts early lymphoid development, we have investigated lineage restriction events within the common lymphoid progenitor (CLP) compartment in IL-7 knockout mice. This revealed that although IL-7 deficiency had a minor impact on the development of LY6D− multipotent CLPs, the formation of the lineage restricted LY6D+ CLP population was dramatically reduced. This was reflected in a low-level transcription of B-lineage genes as well as in a loss of functional B-cell commitment. The few Ly6D+ CLPs developed in the absence of IL-7 displayed increased lineage plasticity and low expression of Ebf-1. Absence of Ebf-1 could be linked to increased plasticity because even though Ly6D+ cells develop in Ebf-1–deficient mice, these cells retain both natural killer and dendritic cell potential. This reveals that IL-7 is essential for normal development of Ly6D+ CLPs and that Ebf-1 is crucial for lineage restriction in early lymphoid progenitors.
Introduction
The development of B-lymphoid cells from hematopoietic stem cells in the bone marrow (BM) is achieved through a stepwise process where multipotent cells gradually lose lineage potentials before committing to B-lineage cell fate. The precise mechanism for this is still under investigation, but several factors with key roles in this process have been identified. One of these is the cytokine IL-7 because in the absence of either the cytokine1 or the α-chain of the receptor,2 B-lymphocyte development is severely impaired and only low numbers of B-lineage progenitors can be found in the BM of adult mice. Even though T-cell development is impaired in the absence of functional IL-7 signaling, the reduction in T-lymphocyte progenitors can be rescued by ectopic expression of Bcl-2.3-5 Expression of this antiapoptotic factor is, however, insufficient to rescue B-lymphocyte development,4,6 suggesting that although IL-7 is permissive for T-cell progenitors, this cytokine might play a more unique role in B-lineage differentiation. Although the block in development of committed CD19+ B-lineage progenitors is obvious, the function of IL-7 signaling in early lymphoid cells is more controversial. Although the early thymic progenitors essentially lack expression of IL-7R,7 the common lymphoid progenitor (CLP) compartment8 as well as a small fraction of the lymphoid-primed multipotent progenitor (LMPP) compartment9 express the receptor. Initial investigations of IL-7R–deficient mice suggested that the CLP compartment developed normally, whereas the development of the progenitor B-cell compartment was dramatically impaired.10 The obvious disadvantage of studying early lymphoid compartments using IL-7R–deficient mice motivated further investigations of the CLP compartment using IL-7–deficient mice where the conventional surface markers for the identification of early populations can be used. These experiments indicated that the addition of IL-7 to CLPs from IL-7–deficient mice was not sufficient to allow for proper B-cell development in vitro, suggesting that IL-7 is critical for the development of B-lineage competent CLPs.11,12 The B-lineage potential could be partially rescued by ectopic expression of the transcription factor Ebf-1, present at reduced levels in CLPs from IL-7–deficient mice compared with their wild-type (wt) counterparts.11-13 Expression of this transcription factor in the CLP compartment is of highest relevance for B-lymphocyte commitment because targeted disruption of the Ebf-1 gene results in that the transcription of B-lineage–associated genes is dramatically reduced.14 The Ebf-1 gene also has been reported to be a direct target for Stat-5, one of the critical signal transducers downstream of the IL-7 receptor in B-cell development,13,15 creating a direct link between IL-7 signaling and Ebf-1 expression.16 Although most of these findings would indicate that IL-7 is critical for normal expression of Ebf-1 and preserved B-lineage potential in B-cell progenitors,13,15 it was reported recently that inactivation of the Stat-5 gene in early lymphoid progenitors did not result in any profound reduction of Ebf-1 expression in early B-cell progenitors.17 Furthermore, the deficiencies in B-cell development could be largely rescued by ectopic expression of Bcl-2,17 arguing for a permissive role of IL-7 in early B-lymphocyte development.
Hence, even though it is obvious that IL-7 is important for B-lymphoid development, the role of this crucial cytokine in lineage decision events remains elusive. In an attempt to clarify the role and functions of IL-7 in early B lymphopoiesis, we have used the recent characterization of functional subpopulations within the CLP compartment18 to extend the phenotypic analysis of IL-7–deficient mice. Even though the CLP has long been treated as a multipotent population, recent findings propose that the lineage negative (Lin−)Flt-3+IL-7R+ScalowKitlow CLP compartment8,19 is composed of at least 3 distinct populations with differences in lineage potential.20 These subpopulations are defined based on the expression of Rag-1 or LY6D,20,21 both serving as markers of progenitor cells lacking natural killer (NK) and dendritic cell (DC) potential as well as on expression of Igll1 (λ5) on Ly6D+ cells marking B-lineage–committed cells.22 Here, we report that in the absence of IL-7, the development of committed B-lymphoid cells within the CLP compartment is severely disrupted. The few residual CLPs that express Ly6D retained NK and DC potential normally lost in these cells. The lineage plasticity can be explained by reduced levels of Ebf-1 in these cells because Ly6D+ CLPs from Ebf-1–deficient mice retained NK and DC potential. Thus, we suggest that IL-7 is critical for normal induction of Ebf-1 expression being crucial for appropriate lineage restriction in the CLP compartment.
Methods
Transgenic mice models
Animal procedures were performed with consent from the local ethics committee at Lund University (Lund, Sweden) and Linköping University (Linköping, Sweden). Bone marrows were harvested from 10- to 15-week-old C57/B6 IL-7−/−,1 C57/B6 IL-7−/− hCD25(λ5)Tghet (hCD25 λ5 promoter transgenic),23 and wt C57/B6 mice. Ebf-1–deficient mice was obtained from a crossing of Ebf-1 C57/B6 to 129 mice24 because the mutation causes embryonic lethality on a pure C57/B6 background.14 All mice used in the experiments are littermates.
FACS staining and purification of bone marrow cells
For analysis and cell sorting of bone marrow cells, FC-blocked (CD16/CD32, 2.4G2) cells were further stained with antibodies against the indicated antigens (see supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). All antibodies were purchased from BioLegend unless otherwise stated. For progenitor isolation, bone marrow cells were subjected to MACS column enrichment of KIT+ cells using anti-CD117 immunomagnetic beads (Miltenyi Biotec) before antibody staining. Analysis and cell sorting was done on an FACSAria cell sorter (BD Biosciences). All gates were put according to fluorescence minus one (FMO) controls.
In vitro evaluation of NK-, B-, and T-cell potential by OP9/OP9DL1 coculture
For evaluation of NK-, B-, and T-cell potential, cells were deposited (using a FACSAria cell sorter) directly into 96-well plates containing preplated (2000 cells/well) OP9/OP9DL cells. NK cultures (on OP9 stroma layers) were supplemented with 20 ng/mL IL-15, 40 ng/mL IL-2, 10 ng/mL KIT ligand (KL), 10 ng/mL Fms-like tyrosine kinase 3 ligand (FLT3L), and 10 ng/mL IL-7. B cultures (on OP9 stroma layers) were supplemented with 10 ng/mL KL, 10 ng/mL FLT3L, and 10 ng/mL IL-7. T cultures (on OP9DL stroma layers) were supplemented with 10 ng/mL KL, 10 ng/mL FLT3L, and 10 ng/mL IL-7. All cytokines were acquired from PeproTech. Cultures were substituted with fresh cytokines every 7 days. Opti-MEM (Invitrogen) supplemented with 10% FCS, 50 μg/mL gentamicin, and 50μM β-mercaptoethanol was used for maintaining the OP9/OP9DL stroma cell lines as well as for the cocultures.
Cocultures were evaluated by FACS staining with CD19 (1D3), B220 (RA3-6B2), and propidium iodide (PI) for B OP9 cocultures; CD19 (1D3), NK1.1 (PK136), and PI for NK OP9 cocultures; and CD19 (1D3), CD90.2/Thy1.2 (53-2.1), and PI for T OP9DL cocultures. B OP9 cocultures were evaluated at day 8, whereas NK OP9 and T OP9DL cocultures were evaluated at day 15 or 16. Samples were analyzed on an FACSCalibur or FACSCanto flow cytometry system (BD Biosciences).
In vitro evaluation of myeloid potential
We have modified a previously described method to evaluate myeloid potential.22 In brief, 150 cells were sorted in 3 mL of medium (Opti-MEM supplemented with 10% FCS, 50 μg/mL gentamicin, 50μM β-mercaptoethanol, 25 ng/mL KL, 25 ng/mL FLT3L, 5 ng/mL IL-3, 5 ng/mL human colony-stimulating factor 1 [M-CSF], 5 ng/mL human colony-stimulating factor 2 [GM-CSF], and 10 ng/mL CSF 3), and 20 μL of medium was plated into each well of 2 72-well plates (Nunc MiniTrays; Thermo Fisher Scientific). Wells were scored after 5 days (using an inverted light microscope) for clonal growth and clone size.
Q-RT-PCR
Quantitative RT-PCR analysis of sorted cells was performed as described previously.25 Assays-on-Demand probes used were as follows: Pax5, Mm00435501_m1; Hprt, Mm00446968_m1; Ebf1, Mm00432948_m1; Rag1, Mm01270936_m1; Cd79a (Mb-1), Mm00432423_m1; Notch1, Mm00435245_m1; and Tdt (Dntt), Mm00493500_m1.
Affymetrix gene expression and data analysis
Gene expression analysis of single cells by multiplex RT-PCR
Multiplex single-cell RT-PCR analysis was performed as described previously.22 For primer sequences, please see supplemental Methods.
Results
IL-7 is critical for the normal maintenance of IL-7 receptor-positive multipotent progenitor cells
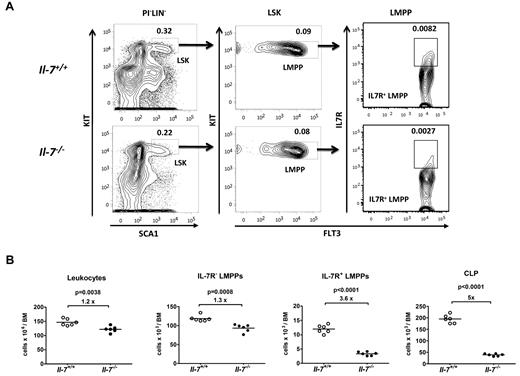
Even though the majority of the IL-7R+ cells in the BM are B-lymphoid–restricted progenitors, the receptor also can be found on noncommitted progenitor cells, suggesting that IL-7 may have effects on the earliest stages of blood cell development. To explore this possibility, we investigated the presence of early hematopoietic progenitor cells in the BM of IL-7–deficient mice. This revealed a small overall reduction (1.2-fold) in total numbers of mononucleated cells in the BM of IL-7–deficient, compared with wt mice (Figure 1A-B). This was comparable to the 1.3-fold reduction observed in the Flt-3+IL-7R−LSK (LMPP) compartment (Figure 1A-B). However, when we analyzed the presence of Flt-3+IL-7R+ LSK cells, this population was reduced 3.6 times (Figure 1A-B), suggesting that IL-7 is involved in the maintenance of the IL-7R+ LMPP compartment. To investigate a potential role for IL-7 signaling in early lymphoid priming, we analyzed the expression levels of the lymphoid associated genes Rag1, Notch1, and TdT (Dntt) in LMPPs from wt and IL-7–deficient mice by Q-PCR (supplemental Figure 1). No major differences in the expression of any of these genes could be detected either by single-cell or Q-PCR, suggesting that IL-7 is not directly involved in lymphoid priming in LMPPs. Subsequent maturation of the LMPPs toward the lymphoid lineage results in CLPs with a lower expression of Kit and Sca-1.8,19 Investigating the absolute numbers of Flt3+IL-7R+KitlowSca-1low CLPs in wt and IL-7–deficient mice revealed that this population was reduced 5-fold in IL-7–deficient mice (Figure 1B). Hence, IL-7 is crucial for the normal maintenance of early IL-7 receptor-expressing cells, but the factor is not essential for early lymphoid priming in the LMPP.
IL-7 is important for the normal maintenance of early IL-7 receptor-positive progenitors. (A) Representative FACS plots showing the gating strategies used to identify IL-7R+ early progenitor cells (LMPPs and CLPs) among lineage low/negative progenitors from normal (IL-7+/+) as well as IL-7–deficient (IL-7−/−) mice, as indicated. The percentages in the gates are mean percentages of total BM-nucleated cells. LIN cocktail contains antibodies against CD11b, GR1, TER119, CD3, CD11c, NK1.1, CD19, and CD45R/B220. (B) Absolute numbers of the total cellularity, IL-7R−LMPPs, IL-7R+LMPPs and CLPs in the BM (femurs, tibias, and crista iliac) of wt and IL-7–deficient mice (n = 6 of each genotype). Each dot represents the number from an individual mouse, and the horizontal line represents the mean in each group. Unpaired t test was used to compare the difference between wt and IL-7−/− mice. Data are collected from 2 independent experiments.
IL-7 is important for the normal maintenance of early IL-7 receptor-positive progenitors. (A) Representative FACS plots showing the gating strategies used to identify IL-7R+ early progenitor cells (LMPPs and CLPs) among lineage low/negative progenitors from normal (IL-7+/+) as well as IL-7–deficient (IL-7−/−) mice, as indicated. The percentages in the gates are mean percentages of total BM-nucleated cells. LIN cocktail contains antibodies against CD11b, GR1, TER119, CD3, CD11c, NK1.1, CD19, and CD45R/B220. (B) Absolute numbers of the total cellularity, IL-7R−LMPPs, IL-7R+LMPPs and CLPs in the BM (femurs, tibias, and crista iliac) of wt and IL-7–deficient mice (n = 6 of each genotype). Each dot represents the number from an individual mouse, and the horizontal line represents the mean in each group. Unpaired t test was used to compare the difference between wt and IL-7−/− mice. Data are collected from 2 independent experiments.
IL-7 is critical for the development of B-lineage–committed progenitor cells
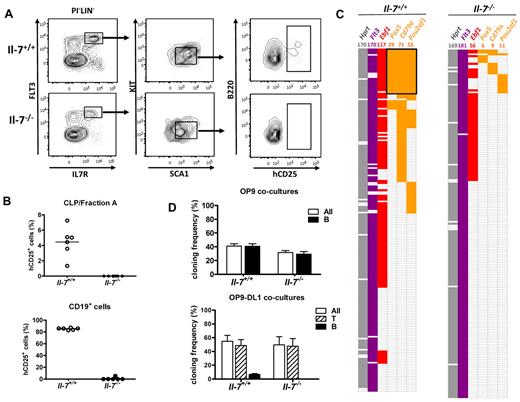
The reduction in these early compartments was, however, not as dramatic as would be predicted from the severe loss of B-cell progenitors observed in IL-7–deficient mice, supporting the notion that IL-7 play a crucial role in the maturation of the earliest B-cell–restricted cells. To investigate this, we crossed IL-7–deficient mice to a strain carrying a reporter transgene where the expression of a human CD25 reporter gene is under the control of the λ5 (Igll1) promoter.23 The expression of this transgene has been reported previously to correlate to B-lineage restriction already in B220−CD19− cells,22 creating a useful tool for investigations of early B-lymphoid commitment. The IL7R+FLT-3+CD19− compartment from wt mice contained ∼ 5% of λ5 reporter-positive cells (Figure 2A-B), whereas essentially no reporter-positive CLPs or B220+ fraction A cells could be detected in the BM from IL-7–deficient mice. Because the expression of the Igll1 reporter is restricted to early B-cell progenitors,23 we were able to investigate the maturation stage of the few CD19+ cells detected in the BM of IL-7–deficient mice. Although 80% of the CD19+ BM cells from a wt mouse expressed the reporter gene, essentially no reporter-positive CD19-expressing cells were detected in BM of IL-7–deficient mice (Figure 2B). This argues for that the CD19+ cells detected in the BM of IL-7–deficient mice represents recirculating mature cells rather than B-lineage progenitors.
CLPs from IL-7 knockout mice are deficient in B-lymphoid commitment but retain their B-lymphoid potential. (A) Representative FACS plots of reporter gene expression within the LIN−Flt-3+IL-7R+Sca1lowKitlow CLP/Fraction A compartments. LIN cocktail contains antibodies against CD11b, GR1, TER119, CD3, CD11c, LY6C, and NK1.1. (B) Summary of the FACS data analyzing reporter gene expression in CLP/Fraction A cells or CD19+ cells collected from 6 IL+/+ and 6 IL−/− mice. Each dot represents one IL+/+ (white) or IL−/− (black) mouse, and the horizontal line represents the mean in each group. (C) Color-coded bar with data collected by single-cell multiplex RT-PCR analysis from CLPs sorted from normal and IL-7–deficient mice. Each horizontal line of boxes represents a single investigated cell collected from 2 independent sorting experiments. A colored box indicates that an RT-PCR product from a given gene could be detected on an ethidium bromide–stained agarose gel. (D) Top: Lymphoid cell cloning frequency of seeded single CLPs collected from normal (IL-7+/+) or IL-7–deficient mice (IL-7−/−; white bars), and the frequency of clones that contain cells expressing CD19 as analyzed by flow cytometry (black bars) after 8 days of incubation on OP9 stroma cells under B-cell–promoting culture conditions. Bottom: Data from cocultures of single sorted wt or IL-7–deficient CLPs incubated on OP9DL stroma cells to stimulate the development of THY1.2+ T-lineage cells. White bars represent total lymphoid cell cloning frequency, striped bars the frequency of T-cell clones, and black bars the frequency of CD19+ B-cell clones. The data are presented as mean + SEM from 3 independent experiments. Cells in both panels have been cultured in the presence of 10 ng/mL KL, 10 ng/mL IL-7, and 10 ng/mL FLT3L.
CLPs from IL-7 knockout mice are deficient in B-lymphoid commitment but retain their B-lymphoid potential. (A) Representative FACS plots of reporter gene expression within the LIN−Flt-3+IL-7R+Sca1lowKitlow CLP/Fraction A compartments. LIN cocktail contains antibodies against CD11b, GR1, TER119, CD3, CD11c, LY6C, and NK1.1. (B) Summary of the FACS data analyzing reporter gene expression in CLP/Fraction A cells or CD19+ cells collected from 6 IL+/+ and 6 IL−/− mice. Each dot represents one IL+/+ (white) or IL−/− (black) mouse, and the horizontal line represents the mean in each group. (C) Color-coded bar with data collected by single-cell multiplex RT-PCR analysis from CLPs sorted from normal and IL-7–deficient mice. Each horizontal line of boxes represents a single investigated cell collected from 2 independent sorting experiments. A colored box indicates that an RT-PCR product from a given gene could be detected on an ethidium bromide–stained agarose gel. (D) Top: Lymphoid cell cloning frequency of seeded single CLPs collected from normal (IL-7+/+) or IL-7–deficient mice (IL-7−/−; white bars), and the frequency of clones that contain cells expressing CD19 as analyzed by flow cytometry (black bars) after 8 days of incubation on OP9 stroma cells under B-cell–promoting culture conditions. Bottom: Data from cocultures of single sorted wt or IL-7–deficient CLPs incubated on OP9DL stroma cells to stimulate the development of THY1.2+ T-lineage cells. White bars represent total lymphoid cell cloning frequency, striped bars the frequency of T-cell clones, and black bars the frequency of CD19+ B-cell clones. The data are presented as mean + SEM from 3 independent experiments. Cells in both panels have been cultured in the presence of 10 ng/mL KL, 10 ng/mL IL-7, and 10 ng/mL FLT3L.
To verify the loss of B-lineage–restricted gene expression in the CLP compartment of IL-7–deficient mice, we performed single-cell multiplex RT-PCR from CLPs collected from wt or IL-7–deficient mice (Figure 2C). This revealed that although 12% of the CLPs obtained from wt mice coordinately expressed the B-lineage genes Pou2af1, CD79α, and Pax5, we were unable to detect coordinated gene expression in any of the 182 investigated CLPs sorted from IL-7–deficient mice. This shows that the normal transcription of B-lineage genes in the CLP compartment is disrupted in the absence of functional IL-7 signaling and supports the idea that this cytokine may be directly involved in the regulation of early B-lineage priming and commitment within the CLP compartment. To extend this analysis, we decided to investigate the developmental potential of CLPs at the single-cell level. To this end, we sorted CLPs from wt or IL-7–deficient mice and cultured them on either OP9 cells, supporting high level of B-cell differentiation, or OP9 cells expressing δ ligand (OP9DL), creating conditions stimulating T-cell development from uncommitted progenitors. The cells were seeded as single cells and incubated in the presence of KL, FL, and IL-7 for 8 days to generate B-lineage cells and for 16 days for the development of T-lineage cells. After the culture period, the content of each well was analyzed for expression of CD19 and Thy1.2 by flow cytometry. Seeding wt CLPs on OP9 cells resulted in that CD19+ cells could be generated from 40% of the wt cells, whereas CLPs from IL-7–deficient mice generated CD19+ cells at a frequency of 35% (Figure 2D). This suggests that CLPs from IL-7–deficient mice possess a robust B-lineage potential that can be rescued by the addition of exogenous IL-7. On incubation of the CLPs on OP9DL cells, the majority of the wt CLPs developed into T-lineage cells; however, ∼ 10% of the clones consisted of CD19+ cells (Figure 2D). This is a highly reproducible finding in agreement with the idea that a fraction of the classic CLPs are B-lineage committed.20,22 Seeding CLPs from IL-7–deficient mice on OP9DL cells resulted in that all the progenitors generated T-cell clones (Figure 2D), providing functional evidence for that IL-7 is critical for the development of B-lineage–restricted progenitors.
IL-7 is critical for the development of LY6D+-lineage–restricted CLPs
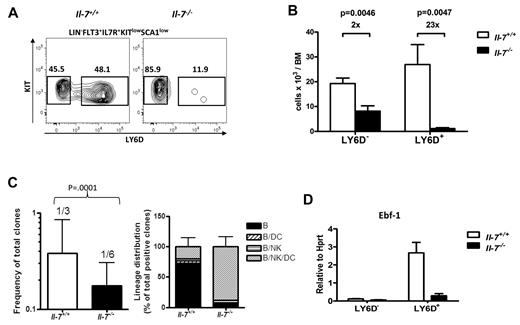
Having defined that the absence of IL-7 results in a developmental block within the CLP compartment, we sorted cells from wt and IL-7–deficient mice and performed global gene expression analysis. Comparing the gene expression pattern obtained to that observed for CLPs from wt mice supported that these cells displayed reduced expression of B-lineage–restricted genes, including CD79, Pou2af1, Igll1, Pax-5, and VpreB1 (supplemental Figure 2). However, we also detected reduced expression of Rag-1 and Ebf-1, both reported to be up-regulated already in the transition from Ly6D− multipotent CLPs into the more lineage-restricted Ly6D+ stage of development.20,21 Comparing the obtained gene expression pattern from CLPs from IL-7–deficient mice to that of LY6D− and Ly6D+ CLP populations from wt mice demonstrated that their gene expression profile was rather similar to that of the Ly6D− cells, suggesting a developmental block already in the Ly6D− to Ly6D+ transition in the absence of IL-7. To investigate this possibility, we analyzed the presence of Ly6D+ CLPs in wt and IL-7–deficient mice. This revealed that although the reduction in absolute numbers of Ly6D− cells was ∼ 2-fold in the IL-7–deficient mice, the difference in Ly6D+ cells was 23-fold, supporting the idea that IL-7 is critical for the development of the B/T-lineage–restricted CLPs (Figure 3A-B). To investigate the lymphoid potential of the Ly6D+ and Ly6D− cells from the IL-7–deficient mice, we seeded single cells on OP9 stroma cells and added cytokines, allowing both B and NK cell development. This revealed that the Ly6D− cells from the IL-7–deficient mice generated both B (CD19+) with normal expression levels of Ebf-1, Pax-5, and λ5 (supplemental Figure 3B) and NK (NK1.1+) cells to a level comparable to that of wt mice, supporting the idea that these early CLPs are functional with regard to the ability to generate lymphoid cells (supplemental Figure 3A). To investigate whether IL-7 is involved in the reduction of myeloid potential observed in the LMPP to CLP transition, we cultured single LMPPs or Ly6D− CLPs from wt and IL-7–deficient mice in Terasaki plates under conditions highly permissive for the development of myeloid cells. Although ∼ 50% of the LMPPs from wt or IL-7–deficient mice generated clones covering > 50% of the well after 5 days, CLPs from either of the mice generated small clones with less than 50 cells (supplemental Figure 3C). Hence, IL-7 is not critical for the reduction in myeloid potential observed in the transition from LMPPs to CLPs, suggesting that the earliest events in lymphoid lineage restriction occur independently of IL-7. To investigate the lineage potentials of the Ly6D− cells in vivo, we transplanted sorted CD45.2+Ly6D− CLPs from wt and IL-7–deficient CD45.2 mice into irradiated CD45.1 recipient mice and compared the generation of B, NK, T, DC, and myeloid cells in spleen and T-lineage cells in thymus, 13 days after transplantation (supplemental Figure 4). This revealed that the cells generated the different lineages in a comparable manner independently of whether they had been exposed to IL-7 or not during their maturation, supporting the notion that the earliest stages of CLP development and myeloid lineage restriction are not critically dependent on IL-7. To obtain a molecular profile of these cells, we performed gene expression microarray analysis of Ly6D− CLPs from wt or IL-7–deficient mice and Ly6D+ cells from wt mice (supplemental Figure 5). This revealed that although there were clear differences in gene expression patterns between the Ly6D+ and Ly6D− CLPs from wt mice, the Ly6D− cells from the IL-7–deficient mice displayed a highly similar gene expression pattern to their counterparts from wt mice. Hence, although IL-7 is crucial for driving the maturation of the Ly6D+ CLP compartment, this cytokine seems less important for the development of Ly6D− CLPs.
B-lymphoid development is blocked in the transition from LY6D− to LY6D+ CLP populations. (A) Representative FACS analysis displaying LY6D+ and LY6D− populations within the Flt-3+IL-7R+Sca1lowKitlow CLP from BM (femurs and tibias) of normal (IL-7+/+) and IL-7–deficient (IL-7−/−) mice. The numbers in the panels are mean percentages of LY6D+/− cells of total CLP. (B) Collective data displaying absolute numbers of LY6D− and LY6D+ CLP populations in wt (white bars) and IL-7–deficient mice (black bars). The data are mean + SEM, from 6 mice of each genotype, analyzed in 2 independent experiments. (C) Total cloning frequency (left) and lineage distribution (right) within BM CLPLY6D+ cells from IL-7+/+ and IL-7−/− mice. The CLPLY6D+ cells were sorted and manually plated at limiting doses (1, 2, and 5 cells/well for IL-7+/+ and 2, 5, and 10 cells per well for IL-7−/− cells) in 96-well plates. The frequencies were calculated based on numbers of positive wells using L-Calc software (StemCell Technologies, http://www.stemcell.com/tutorials/lcsetup.exe). The bars show mean 95% confidence interval. The difference between the 2 genotypes was compared by 2-tailed t test built in L-Calc software. For investigation of lineage potentials, the obtained cultures were analyzed by FACS for expressions of different lineage markers, including B cells (CD19+B220+), DCs (CD11C+), and NK cells (NK1.1+). The clones were defined as positive when having a minimum of 0.02% of each of the B, NK, and DC lineages and ≥ 10 gated events with an analysis of a minimum 25 000 blood cells. Bars are mean + SD. (D) Q-PCR analysis of expression of Ebf-1 in LY6D+ and LY6D− CLPs from normal (IL-7+/+) and IL-7–deficient (IL-7−/−) mice. The data are normalized to the expression of Hprt, collected from 3 independent sorting experiments, and analyzed by triplicate Q-PCR reactions. The data are mean + SEM.
B-lymphoid development is blocked in the transition from LY6D− to LY6D+ CLP populations. (A) Representative FACS analysis displaying LY6D+ and LY6D− populations within the Flt-3+IL-7R+Sca1lowKitlow CLP from BM (femurs and tibias) of normal (IL-7+/+) and IL-7–deficient (IL-7−/−) mice. The numbers in the panels are mean percentages of LY6D+/− cells of total CLP. (B) Collective data displaying absolute numbers of LY6D− and LY6D+ CLP populations in wt (white bars) and IL-7–deficient mice (black bars). The data are mean + SEM, from 6 mice of each genotype, analyzed in 2 independent experiments. (C) Total cloning frequency (left) and lineage distribution (right) within BM CLPLY6D+ cells from IL-7+/+ and IL-7−/− mice. The CLPLY6D+ cells were sorted and manually plated at limiting doses (1, 2, and 5 cells/well for IL-7+/+ and 2, 5, and 10 cells per well for IL-7−/− cells) in 96-well plates. The frequencies were calculated based on numbers of positive wells using L-Calc software (StemCell Technologies, http://www.stemcell.com/tutorials/lcsetup.exe). The bars show mean 95% confidence interval. The difference between the 2 genotypes was compared by 2-tailed t test built in L-Calc software. For investigation of lineage potentials, the obtained cultures were analyzed by FACS for expressions of different lineage markers, including B cells (CD19+B220+), DCs (CD11C+), and NK cells (NK1.1+). The clones were defined as positive when having a minimum of 0.02% of each of the B, NK, and DC lineages and ≥ 10 gated events with an analysis of a minimum 25 000 blood cells. Bars are mean + SD. (D) Q-PCR analysis of expression of Ebf-1 in LY6D+ and LY6D− CLPs from normal (IL-7+/+) and IL-7–deficient (IL-7−/−) mice. The data are normalized to the expression of Hprt, collected from 3 independent sorting experiments, and analyzed by triplicate Q-PCR reactions. The data are mean + SEM.
One potential explanation to the loss of cells could be decreased expansion of the Ly6D+ CLPs; however, cell cycle analysis of these early compartments did not support this hypothesis because the Ly6D+ cells from IL-7–deficient mice rather contained a larger fraction of cells displaying the signs of being in cycle than the corresponding cells from wt mice (supplemental Figure 6).
Although the transition from the LMPP to the Ly6D− compartment is associated with a reduction in myeloid potential, the maturation into Ly6D+ cells is linked to a reduction in NK and DC potential.20,21 To investigate whether these lineage restriction events were dependent of IL-7, we cultured Ly6D+ CLPs from wt and IL-7–deficient mice with OP9 stroma cells in the presence of IL-2 and IL-15 in addition to FL, KL, and IL-7 used for B-cell cultures, creating conditions allowing for combined development of NK/B and DC lineage cells. Because our initial single-cell analysis suggested there was a difference in cloning efficiency between Ly6D+ CLPs from wt or IL-7–deficient mice, we performed limited dilution experiments by seeding 1, 2, 5, and 10 cells on OP9 stroma cells. The cloning frequency was subsequently calculated based on the numbers of positive wells using the L-Calc software (StemCell Technologies, http://www.stemcell.com/tutorials/lcsetup.exe). This revealed that although our experimental conditions allowed clone development from 1 of 3 seeded cells from wt mice, only 1 of 6 Ly6D+ cells from IL-7–deficient mice were able to generate a clone under these conditions (Figure 3C). The functional differences became even more apparent when we investigated the cellular content of the clones generated because although the majority of the clones generated from wt CLPs contained only CD19+ cells, the opposite was true for the Ly6D+ CLPs from IL-7–deficient mice because a majority of the Ly6D+ cells gave rise to mixed colonies with B cells, NK cells, DCs, and progeny (Figure 3C; supplemental Figure 7). This reveals that Ly6D+ CLPs generated in the absence of IL-7 maintain in vitro B-lineage potential but are not properly lineage restricted.
One potential explanation to the lack of proper lineage restriction in the Ly6D+ cells from IL-7–deficient mice would be reduced expression of lineage restricting transcription factors. Even though Pax-5 has been suggested to be the key regulator of B-cell commitment, its has been proposed that Ebf-1 expression is directly regulated by IL-7 signaling16 and that ectopic expression of Ebf-1 is sufficient to cause lineage restriction in Pax-5–deficient progenitors.28 Furthermore, although the expression of Pax-5 can only be detected in a fraction of the CLPs,22,29 Ebf-1 expression is more abundant and dramatically increased in the transition from Ly6D− to Ly6D+ cells.21,22 It has been reported that Ebf-1 expression is reduced in IL-7–deficient mice;11-13 however, in light of our data, this could be explained by the disruption in CLP development because the cells expressing high levels of Ebf-1 are dramatically reduced. Hence, we investigated the expression of Ebf-1 message in Ly6D− and Ly6D+ CLPs from wt and IL-7–deficient mice by Q-PCR. This suggested that the expression of Ebf-1 was reduced in Ly6D+ cells developed in the absence of IL-7 (Figure 3D), supporting the idea that IL-7 signaling is crucial for the normal induction of Ebf-1 expression in the CLP compartment.
Ebf-1 is crucial for the repression of NK and DC potential in Ly6D+ CLPs
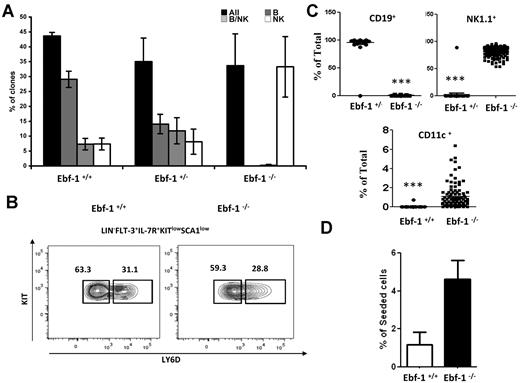
Knowing that Ly6D+ cells developed in the absence of IL-7 display enhanced lineage plasticity and express reduced amounts of Ebf-1, we wanted to investigate a potential direct role for Ebf-1 in lineage restriction in the CLP compartment. To this end, we seeded single CLPs from wt, Ebf-1 heterozygote, and Ebf-1–deficient mice24 and investigated their ability to develop into B and NK cells in vitro. Cells from all the different genotypes generated clones with a comparable frequency (Figure 4A). However, although the majority of the wt cells generated clones containing only CD19+ cells, this fraction was reduced in the heterozygote Ebf-1 mutant and because of the crucial role for Ebf-1 in B-cell development,24 absent when we used cells from Ebf-1–deficient mice. However, the finding that all the Ebf-1–deficient CLPs had the potential to develop into NK1.1+ cells suggested that no B-lineage–restricted CLPs developed in these mice. This could either be explained by that Ebf-1 is crucial for the development of the Ly6D+ compartment or that Ly6D+ cells are plastic unless they are capable to up-regulate the expression of Ebf-1. To resolve this issue, we analyzed the composition of the CLP compartment in 5- to 6-week-old Ebf-1–deficient mice, an age where the functional defects of Stat-5 deletion are apparent.17 In contrast to the IL-7–deficient mice, mice lacking Ebf-1 display a normal distribution between Ly6D− and Ly6D+ cells (Figure 4B). This revealed that IL-7 signaling is not dependent on Ebf-1 and positions this transcription factor downstream of IL-7 in the developmental hierarchy. However, cultivation of single Ly6D+ CLPs under conditions allowing for NK, B, and DC development revealed that even though both wt and Ebf-1–deficient Ly6D+ CLPs generated clones with comparable efficiency, the cells sorted from wt littermates almost exclusively gave rise to CD19+ clones, whereas cells sorted from Ebf-1–deficient mice developed into both NK1.1 and CD11c positive cells with a high frequency (Figure 4C). To investigate changes in myeloid potential, we cultured Ly6D+ CLPs under myeloid conditions and investigated the frequency of clones obtained after 5 days of incubation (Figure 4D). This revealed that even though the overall cloning efficiency was low as expected from Ly6D+ CLPs, Ebf-1–deficient cells generated more myeloid clones than wt cells. This suggests that Ebf-1 is crucial for normal lineage restriction events in the CLP compartment and supports the idea that Ebf-1 mediates IL-7–dependent lineage restriction.
Ebf-1 is critical for IL-7 mediated lineage restriction in the CLP compartment. (A) Diagrams with the overall cloning frequency (black bars) and the development of CD19+ (gray bars), mixed lineages (CD19+ cells and NK1.1+ cells (striped bars), or NK1.1+ (white bars) from single wt (Ebf-1+/+) Ebf-1 heterozygote (Ebf-1+/−) or Ebf-1–deficient (Ebf-1−/−) CLPs as indicated. The data are collected from 3 independent experiments using 5- to 6-week-old Ebf-1–deficient mice, and the data are mean + SEM. (B) Representative FACS analysis displaying LY6D+ and LY6D− populations within the Flt-3+IL-7R+Sca1lowKitlow CLP from BM (femurs and tibias) of normal (Ebf-1+/+) and Ebf-1–deficient (Ebf-1−/−) mice. The numbers in the plots are mean percentages of LY6D+/− cells of total CLP analyzed in 2 independent experiments. (C) Diagrams with the percentage of CD19+, NK1.1+, or CD11c+ cells obtained in culture after seeding single Ly6D+ CLPs from wt (Ebf-1+/+) or Ebf-1–deficient (Ebf-1−/−) mice. Each dot represents a single clone. The difference between 2 genotypes was evaluated by 2-tailed t test. The graph in panel D displays the cloning frequency of wt and Ebf-1–deficient Ly6D+ CLPs after 5 days of incubation under myeloid conditions. The data are collected from 288 seeded Ebf-1–deficient and 462 wt cells collected from 2 independent experiments. The error bars indicate SD.
Ebf-1 is critical for IL-7 mediated lineage restriction in the CLP compartment. (A) Diagrams with the overall cloning frequency (black bars) and the development of CD19+ (gray bars), mixed lineages (CD19+ cells and NK1.1+ cells (striped bars), or NK1.1+ (white bars) from single wt (Ebf-1+/+) Ebf-1 heterozygote (Ebf-1+/−) or Ebf-1–deficient (Ebf-1−/−) CLPs as indicated. The data are collected from 3 independent experiments using 5- to 6-week-old Ebf-1–deficient mice, and the data are mean + SEM. (B) Representative FACS analysis displaying LY6D+ and LY6D− populations within the Flt-3+IL-7R+Sca1lowKitlow CLP from BM (femurs and tibias) of normal (Ebf-1+/+) and Ebf-1–deficient (Ebf-1−/−) mice. The numbers in the plots are mean percentages of LY6D+/− cells of total CLP analyzed in 2 independent experiments. (C) Diagrams with the percentage of CD19+, NK1.1+, or CD11c+ cells obtained in culture after seeding single Ly6D+ CLPs from wt (Ebf-1+/+) or Ebf-1–deficient (Ebf-1−/−) mice. Each dot represents a single clone. The difference between 2 genotypes was evaluated by 2-tailed t test. The graph in panel D displays the cloning frequency of wt and Ebf-1–deficient Ly6D+ CLPs after 5 days of incubation under myeloid conditions. The data are collected from 288 seeded Ebf-1–deficient and 462 wt cells collected from 2 independent experiments. The error bars indicate SD.
Discussion
Even though it has been apparent that IL-7 plays a crucial role in lymphocyte development, different experimental approaches have produced apparently contradictory results that largely can be explained by the finding that IL-7 deficiency blocks the maturation of CLPs. Even though B220+CD43+ pre-pro-B cells develop in the adult IL-7–deficient mice, they lack transcription of B-lineage genes and do not display any ability to develop into CD19+ cells.12 This compartment of cells is highly heterogeneous in a wt mouse and in mice lacking factors crucial for development of early B-cell progenitors an apparently normal B220+CD43+ pre-pro-B-cell compartment may well be completely disrupted with regard to B-lineage progenitors. One striking example is the Ebf-1–deficient mice in which a clear B220+CD43+ population that lacks all B-lymphoid potential in vitro and in vivo, immunoglobulin recombination and transcription of B-lineage genes can be detected.14,24 Rather, we believe that our data suggest that IL-7 is crucial for the proper establishment of the genetic network that drives B-cell commitment already in the CLP compartment. Even though a permissive action of IL-7 in this transition should not be excluded, the finding that ectopic expression of constitutively active Stat-5 causes increased B-cell development in the thymus30 and poor recovery of B-lymphoid cells on ectopic expression of Bcl-23,5,6,28 supports the idea that IL-7 has an instructive role in early B-cell development. It was recently reported that defects in B-cell differentiation after conditional deletion of Stat-5 could be rescued by ectopic expression of antiapoptotic factors.17 However, because these experiments were conducted using a Rag-1–driven Cre-recombinase, we believe that these data can be explained by that Stat-5 is functional in the majority of the LY6D−Raglow/− cells and that the deletion of the gene does not occur until after the cells have reached the IL-7–dependent LY6D+Rag1high stage of development where Ebf-1 expression is already established. The disruption in CLP maturation observed also explains the discrepancies reported regarding the B-lymphoid potential of progenitor cells generated in the absence of IL-7.11,12 Even though the early CLP population found in IL-7–deficient mice has B-cell potential, these cells have several other cell fates open and they also generate B-lymphoid cells with a lower frequency and slower kinetics than the more mature LY6D+Rag1+ cells that compose a large part of the CLPs in a wt mouse.20,21
In addition to providing new insight into the processes that critically depend on IL-7, our analysis also provides information about IL-7–independent processes in early hematopoiesis. The lack of IL-7 causes a small decrease in the overall numbers of mononucleated cells in the BM. Even if this could be a consequence of reduced numbers of lymphoid progenitors, the finding that IL-7R–negative LMPPs display the same reduction could indicate that disruptions in the BM microenvironment may have an impact also on progenitor cells not directly responding to IL-7. The decrease was, however, not as dramatic as that observed for IL-7R–expressing LMPPs. Even though IL-7 is required for maintenance of normal numbers of IL-7R–expressing LMPPs, we observed normal expression of lymphoid associated genes in the LMPP compartment of IL-7–deficient mice. This together with our data supporting normal gene expression and function of the Ly6D− CLPs from IL-7–deficient mice argues against a role for IL-7 in lymphoid lineage priming in the LMPPs. Even though we could detect a reduction in the numbers of thymic NK cells, as reported previously,31,32 we could not detect any reductions in NK cell numbers either in the spleen or in the BM of IL-7–deficient mice (data not shown). This supports the idea that NK cell development is normal or, because there is a reduction in the LY6D− compartment, possibly enhanced on a per cell basis in the absence of IL-7. The idea of an early separation of B and NK cell potential is also supported from lineage tracing experiments using a Rag-1 promoter–driven CRE to induce expression of red fluorescent protein because few reporter-positive NK cells were detected.33
Even though we are unable to draw conclusions about the lineage potential of the Ly6D+ cells in vivo, our in vitro analysis strongly suggest that they are not properly lineage restricted unless exposed to IL-7. Our finding that Ebf-1 is crucial for IL-7–mediated lineage restriction events does not resolve if Ebf-1 alone is sufficient to restrict lineage potential in the absence of functional IL-7 signaling. However, the finding that ectopic expression of Ebf-1 is sufficient to restrict cell fate options after transplantation,28,34 shows that Ebf-1 is a powerful mediator of lineage restriction in the hematopoietic system. Even though the genetic network downstream of Ebf-1 is being unraveled, revealing both repressed and activated target genes,35,36 the temporal regulation of these genes and their function in lineage restriction events remains to be resolved. Our data also reveal that even though functionally impaired, we do find a Ly6D+ CLP compartment in Ebf-1–deficient mice, revealing that IL-7 acts upstream of Ebf-1 and that the initial signaling events as well as myeloid lineage restriction are not critically dependent on this transcription factor. One critical factor in this context is Miz-1 that seems critical for B-cell development and proper IL-7 signaling.37 In the absence of Miz-1, B-cell development in vitro could only be rescued by combined ectopic expression of Ebf-1 and Bcl-2,37 supporting the idea that IL-7 has both instructive and permissive functions in lymphoid development. In addition, to direct effects via induction of antiapoptotic factors, it has been shown that IL-7 modulates metabolism in lymphoid cells. Stat-5 signaling can modulate glucose uptake38 and inactivation of functional IL-7 signaling in mature T cells cause an impairment in glycolytic flux.39 Even though these effects have been investigated mainly in T-lineage cells, the large impact on cell function posed by metabolic control motivates further analysis of this issue also in the early development of lymphoid cells in the BM.
In all, we believe our data suggest that IL-7 plays a critical instructive role in B-lineage commitment and even though this cytokine seems to be involved in the maintenance of the most immature IL-7R+ cells, lineage priming, reduction of myeloid potential, and functional activity of these cells remain intact. Development is rather blocked at the defined transition between the multipotent Ly6D− stage and the more restricted Ly6D+ stage, where IL-7 is crucial for the development of properly lineage-restricted cells and the initiation of the B-lymphoid program.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank their colleagues for help with transgenic mice and cell lines.
This study was generously supported by grants from the Swedish Cancer Society, the Swedish Research Council, the Swedish Childhood Cancer Foundation, and the faculty of Medicine at Linköping University.
Authorship
Contribution: P.T., S.Z., J.Å., J.Z., E.W., R.M., J.I.J., and H.Q. designed the research; performed experiments; collected, analyzed, and interpreted data; performed statistical analysis; and wrote the manuscript; and M.S. designed the research, analyzed and interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mikael Sigvardsson, Faculty of Health Sciences, Linköping University Lab1, Level 13, 58185 Linköping, Sweden; e-mail: mikael.sigvardsson@liu.se.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal