Abstract
Diamond-Blackfan anemia (DBA) is a congenital BM failure syndrome characterized by hypoproliferative anemia, associated physical abnormalities, and a predisposition to cancer. Perturbations of the ribosome appear to be critically important in DBA; alterations in 9 different ribosomal protein genes have been identified in multiple unrelated families, along with rarer abnormalities of additional ribosomal proteins. However, at present, only 50% to 60% of patients have an identifiable genetic lesion by ribosomal protein gene sequencing. Using genome-wide single-nucleotide polymorphism array to evaluate for regions of recurrent copy variation, we identified deletions at known DBA-related ribosomal protein gene loci in 17% (9 of 51) of patients without an identifiable mutation, including RPS19, RPS17, RPS26, and RPL35A. No recurrent regions of copy variation at novel loci were identified. Because RPS17 is a duplicated gene with 4 copies in a diploid genome, we demonstrate haploinsufficient RPS17 expression and a small subunit ribosomal RNA processing abnormality in patients harboring RPS17 deletions. Finally, we report the novel identification of variable mosaic loss involving known DBA gene regions in 3 patients from 2 kindreds. These data suggest that ribosomal protein gene deletion is more common than previously suspected and should be considered a component of the initial genetic evaluation in cases of suspected DBA.
Introduction
Since the initial descriptions of heterozygous RPS19 mutations in a subset of Diamond-Blackfan anemia (DBA) patients, significant progress has been made over the past decade in further elucidating the genetic cause of DBA.1 With increasing focus on the ribosomal protein (r-protein) genes, the search for DBA-related genes, initially based on classic genetics techniques including cloning of cytogenetic abnormalities and extended linkage analysis, has shifted to targeted resequencing of the known r-protein genes. Such studies have identified both large and small subunit r-protein gene abnormalities, currently including RPL5, RPL11, RPL35A, RPS7, RPS10, RPS17, RPS19, RPS24, and RPS26 reported as mutated in multiple families and in multiple studies.2-6 Alterations of an even larger number of r-proteins have been identified in isolated patients or families, including RPL3, RPL7, RPL9, RPL14, RPL19, RPL23A, RPL26, RPL35, RPL36, RPS8, RPS15, and RPS27A.5-8 Alhough the significance of these rarer r-protein gene alterations is not yet clear, it is conceivable that many are pathogenic. However, despite the large number of potential DBA genes, sequencing studies evaluating the r-protein gene complement in DBA patients have failed to find a mutation in 40% to 45% of patients.6,7
Dominant-negative effects of a mutant r-protein, particularly in the case of RPS19, have been implicated by some missense coding mutations and demonstrated in model systems.9-11 However, the majority of missense mutations identified in DBA r-proteins are thought to be loss of function. Furthermore, the majority of DBA mutations in RPS19 and the other genes identified are nonsense,12,13 suggesting that allelic haploinsufficiency is sufficient to lead to DBA.14-16 Genomic deletions or rearrangements at r-protein loci may also lead to allelic haploinsufficiency and have been sporadically reported for several r-protein genes.4,17,18
Several strategies are used for the detection of copy number variants (CNVs) in the genome, including focused PCR-based techniques such as multiplex ligation-dependent probe amplification, hybridization microarrays such as array comparative genomic hybridization (aCGH) and single-nucleotide polymorphism genotyping arrays (SNP-array), and next-generation sequencing-based approaches. SNP-array, an increasingly available and cost-effective method for whole-genome CNV detection, is a hybridization-based technology that uses signal intensity data at SNP probes to derive copy number estimates based both on the normalized signal intensities and allele frequencies at a given region. In contrast to aCGH, SNP-array can also detect mosaic copy gain and loss.19 Because most of the Sanger-based resequencing studies used to screen for DBA-related mutations yield sequence data without copy number information, we hypothesized that a significant proportion of those DBA patients whose molecular abnormality remains unidentified may harbor genomic rearrangements or deletions that disrupt r-protein genes or other regions critical in DBA. To test this hypothesis, we screened patients lacking r-protein gene mutations from the Diamond-Blackfan Anemia Registry (DBAR) by SNP-array.20,21
Methods
Patient samples
Fifty-one affected probands, 1 affected sibling, 1 unaffected sibling, and 15 parents without known r-protein gene mutations were studied. Informed consent was obtained from all individuals or guardians through the DBAR in accordance with the Declaration of Helsinki. This study was approved by the institutional review boards of all participating institutions. Forty-one probands had previously been screened for r-protein gene mutations through the DBAR r-protein resequencing study. The remaining 10 patients underwent clinical mutation testing for the 9 common r-protein gene mutations. For SNP array and CGH studies, DNA was isolated after RBC lysis from whole peripheral blood nucleated cells using standard techniques. For lineage-specific SNP array, lymphocytes were isolated using magnetic beads to CD3 and CD19; more than 95% of isolated cells were CD4/8+ or CD20+. The residual cell fraction was considered to be myeloid and was > 98% depleted of CD4/8+ and CD20+ cells.
SNP-array genotyping
Genomic DNA (300 ng) was prepared for analysis using the Infinium HD assay protocol according to the manufacturer's instructions (Illumina).22 Samples were hybridized to Illumina BeadChips, with 24 samples initially run on HumanOmni1-Quad chips and 44 additional samples run on HumanOmniExpress chips. Image data were scanned with a BeadArray reader and intensity and genotype data were extracted using the GenomeStudio (Version 2010.3) genotyping module using cluster definitions provided by the manufacturer (HumanOmni1-Quad) or developed from a 431-patient set evaluated through the National Institutes of Health Undiagnosed Diseases Program (HumanOmniExpress). Call rates were > 98.9% in all samples using a GenCall threshold of 0.15. Normalized signal intensity ratios and B-allele frequency data were exported for CNV analysis. Array data were deposited in the National Center for Biotechnology Information (NCBI)/Gene Expression Omnibus database under accession number GSE31575.
SNP-array CNV detection
Regions of autosomal CNV were identified with PennCNV,23 a hidden Markov model-based CNV-calling algorithm that incorporates both signal intensity and allele frequency data for variant identification using population B-allele frequency data provided by the algorithm developer (HumanOmni1-Quad) or derived from a 431-patient set evaluated through the National Institutes of Health Undiagnosed Diseases Program (HumanOmniExpress), a trained hidden Markov model file provided by the algorithm developer, and a minimum of 10 contiguous SNP parameter for CNV calling. Regions of CNV were annotated for coding genes contained within or 10 kb adjacent to CNV regions by comparison with hg18 UCSC known Gene and NCBI refGene tables. To identify CNV at potentially novel r-protein and established DBA loci, genes in each raw CNV call were queried for the presence of any annotated r-protein gene. To detect novel non-r-protein loci, called CNVs were subsequently filtered to exclude those overlapping with regions reported in the Database of Genomic Variants and tested for the presence of common CNVs in 2 or more DBA probands.24
Quantification of mosaic copy loss
The continuous distribution function of heterozygous B-allele frequency (BAF) data was formed from the selected regions of proband data. Data from the heterozygous BAFs of 5 normal controls were averaged and then randomly dichotomized so that half of the normal data were shifted up and half down by a value predicted from a starting estimate of the degree of mosaicism (f) using a model of mosaic monosomy/disomy. A continuous distribution function of the normal, shifted data was re-formed and subtracted from unknown proband data. This f was iteratively regressed against the proband data until solution of a minimum residual. The final f was taken as the degree of mosaicism in the unknown sample (T.C.M., H.C.D., M. Sincan, D. Adams, D.M.B., J.E.F., A.V., J.M.L., A. Auerbach, E. Ostrander, S. Chandrasekharappa, C. Boerkoel, W. Gahl, Detection and Sensitive Quantification of Mosaicism Using High-Density SNP Arrays and the Cumulative Distribution Function, manuscript submitted).
aCGH
For aCGH, 500 ng of test DNA and a reference male/female DNA mixture (Promega) were labeled with Cy3 or Cy5, respectively, using the NimbleGen labeling kit. The labeled DNAs were hybridized to NimbleGen Human CGH 3 × 720K Whole-Genome Exon-Focused Arrays for 70 hours at 42°C, washed, and then scanned on an Agilent 2 micron scanner (Agilent Technologies). Feature extraction, primary data analysis, and visualization were performed with DEVA Version 1.0.2 software (Roche NimbleGen). For CNV calling, normalized intensity ratios were imported in Partek GS and detected using the genomic segmentation algorithm using a minimum of 10 genomic markers, a P value threshold of 10−3, and a signal-to-noise ratio of 0.3. Array data were deposited in the NCBI/Gene Expression Omnibus database under accession number GSE31575.
Reverse transcription and quantitative PCR
RNA and DNA were isolated from peripheral blood mononuclear cells after separation of whole blood by Ficoll-Paque PLUS (GE Health Sciences) and expansion for 1-5 days in RPMI medium supplemented with 10% FBS, 2mM l-glutamine, and 5 ng/mL of concanavalin A (Sigma-Aldrich) with the AllPrep Mini Kit (QIAGEN), and quantified by fluorescence using the Qubit RNA Assay or the dsDNA HS Assay (Invitrogen). RNA was reverse-transcribed using the iScript cDNA Synthesis Kit (Bio-Rad) after normalization of input RNA in a 20-μL reaction volume following the manufacturer's instructions, and diluted 1:4 after heat inactivation. Quantitative PCR (qPCR) was performed in triplicate for each reaction in a 20-μL reaction volume of 1× SsoFast EvaGreen Supermix (Bio-Rad) and 500nM primers with 40 ng of gDNA or 1 μL of diluted cDNA reaction mixture on a Bio-Rad CFX96 instrument with initial denature of 98°C for 5 minutes, 40 cycles of 98°C for 5 seconds, and 60°C for 5 seconds, followed by melt-curve analysis for reaction specificity. All PCR reactions were linear over 3 orders of magnitude bracketing the experimental results and were > 95% efficient. For quantification of cDNA, RPS17 message was normalized to expression of RPL35A, a large ribosomal subunit mRNA. For genomic copy number, samples were normalized to an intergenic region in the β-globin locus. The primer sequences (forward/reverse) used were RPS17 cDNA: AAGCTCCGCAACAAGATAGC/TCCTGATCCAAGGCTGAGAC, RPL35A cDNA: TGCTGGGAACG- GGACTTCTA/CTGTGTGCTCCCTTTGGTTC, RPS17 gDNA: CAGCCCAGGATGTCTACGTT/ACCCAATGTACCATGCCATT, and β-globin intergenic DNA: GCAAGATGTTGGCCCTAAAA/CAACAAGGTGCC- AAGTCTTT.
rRNA processing
Preparation and quantitation of Northern blots with hybridization probes to the 18SE pre-rRNA region was performed as described previously.25
Results
SNP-array identifies RPS19 and RPS26 gene deletions in DBA patients with normal r-protein gene sequence
Two different SNP-array platforms were used to detect CNVs. Twenty-four samples were run on Illumina HumanOmni1-Quad SNP arrays, which interrogate 1.1 million SNPs and nonpolymorphic CNV loci with a median marker spacing of 1.25 kb. An average of 260 autosomal CNVs were identified per sample (range 166-377), with a median size of 2.8 kb (range, 359 bases to 1.6 Mb). The majority of the variants identified were small, with 80% of CNVs smaller than 10 kb. The remaining 44 samples were run on Illumina HumanOmniExpress arrays, which interrogate 733 thousand SNPs with a median marker spacing of 2.17 kb. On average, 7.4 CNVs were identified per sample (range 1-38) with a median size of 52 kb (range, 2 kb to 1.6 Mb). Compared with the HumanOmni1-Quad chip, the HumanOmniExpress arrays query the majority of informative polymorphic SNP probes of the former, but exclude the nonpolymorphic CNV probes. The difference in average number and size of CNV calls between platforms is largely attributable to increased marker density on the HumanOmni1-Quad chip. Both arrays cover > 95% of the genome, including all annotated r-protein genes, and both have sufficient sensitivity to detect gene-level and larger deletions.
We identified 7 patients with single-copy deletions of known DBA genes after annotation of all r-protein genes within or adjacent to unfiltered CNV regions. We did not identify deletions involving novel r-protein genes from unfiltered CNV calls, nor were we able to identify common regions of CNV outside of known r-protein gene loci after exclusion of the CNV regions reported in the Database of Genomic Variants.24
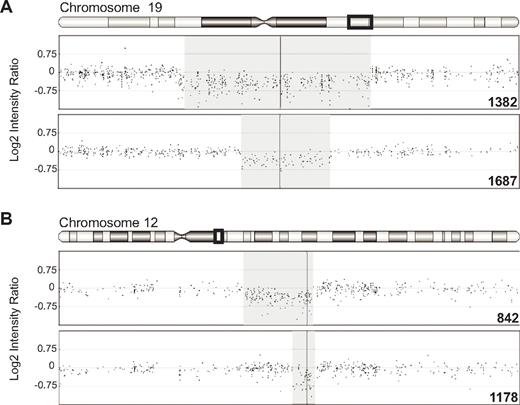
The clinical features and array findings in each patient are outlined in Table 1. Two patients with single-copy deletions of 377 and 828 kb involving RPS19 were identified (Figure 1A). Two additional patients with single-copy deletions of 15 and 85 kb involving RPS26 were identified (Figure 1B). To confirm the SNP-array results, we repeated the analysis with an aCGH platform containing focused probe coverage in annotated exonic regions. The deletions in samples 1178, 1382, and 1687, for which there was sufficient sample for further testing, were confirmed by aCGH with decreased normalized log2 probe intensity in each of the regions identified.
Ribosomal protein gene deletions and associated clinical findings in patients with DBA
| ID . | Region (hg18)* . | Size . | Gene . | Sex . | Age . | Hgb/Hct . | Retic . | eADA . | MCV . | Steroid response . | Physical abnormality . | Other . | Confirmed by aCGH . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1382 | chr19:46620492-47448185 | 828 kb | RPS19 | F | 1 mo | 2/6 | 0.1 | — | — | N | Macrocephaly, developmental delay | MS-BMT at 2.5 y | Yes |
| 1687 | chr19:46898790-47275854 | 377 kb | RPS19 | F | 0 mo | 1.7/5.9 | 0 | — | 95.1 | N | VSD | Yes | |
| 1178 | chr12:54715842-54731191 | 15 kb | RPS26 | M | 3.5 mo | 3.5/9.7 | 0.6 | — | 110 | Y | None | Yes | |
| 842 | chr12:54643476-54728995 | 85 kb | RPS26 | M | 7 mo | 7.2/20.5 | 0.8 | — | 105.8 | Y | None | Remission as teenager | Not tested |
| 886 | chr15:81011018-82589310† | 1.5 Mb | RPS17 | F | 0 mo | 7.6/— | — | — | — | Y | None | Yes | |
| 1314 | chr15:81011018-82623936† | 1.6 Mb | RPS17 | F | 2 mo | 2.6/— | — | ↑ | — | Y | PFO, short stature | Initially steroid responsive, currently transfusion dependent | Not tested |
| 20QL | chr15:81011018-82623936† | 1.6 Mb | RPS17 | M | 2.5 mo | 2.1/— | — | — | — | N | Short stature | Yes | |
| 802 | 3q21-tel | Variable | See¶ | M | 2 y | 7.1/21 | 0.7 | 0.72 | 103 | Y | None | Neutropenia; remission at age 16‡ | N/A |
| 802-2 | 3q21-tel | Variable | See¶ | F | 2 mo | 2.6/7.4 | 0.1 | 1.17 | 90 | N | None | Neutropenia‡ | N/A |
| 80-3§ | — | — | — | F | 2 y | 10.4/ 30.4 | 1.7 | 1 | 93 | Y | None | Neutropenia; remission at age 12‡ | N/A |
| 1786 | 15q | Variable | See# | M | 2 mo | 4.3/12.5 | — | 1.01 | — | Y | None | Remission as teenager | N/A |
| ID . | Region (hg18)* . | Size . | Gene . | Sex . | Age . | Hgb/Hct . | Retic . | eADA . | MCV . | Steroid response . | Physical abnormality . | Other . | Confirmed by aCGH . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1382 | chr19:46620492-47448185 | 828 kb | RPS19 | F | 1 mo | 2/6 | 0.1 | — | — | N | Macrocephaly, developmental delay | MS-BMT at 2.5 y | Yes |
| 1687 | chr19:46898790-47275854 | 377 kb | RPS19 | F | 0 mo | 1.7/5.9 | 0 | — | 95.1 | N | VSD | Yes | |
| 1178 | chr12:54715842-54731191 | 15 kb | RPS26 | M | 3.5 mo | 3.5/9.7 | 0.6 | — | 110 | Y | None | Yes | |
| 842 | chr12:54643476-54728995 | 85 kb | RPS26 | M | 7 mo | 7.2/20.5 | 0.8 | — | 105.8 | Y | None | Remission as teenager | Not tested |
| 886 | chr15:81011018-82589310† | 1.5 Mb | RPS17 | F | 0 mo | 7.6/— | — | — | — | Y | None | Yes | |
| 1314 | chr15:81011018-82623936† | 1.6 Mb | RPS17 | F | 2 mo | 2.6/— | — | ↑ | — | Y | PFO, short stature | Initially steroid responsive, currently transfusion dependent | Not tested |
| 20QL | chr15:81011018-82623936† | 1.6 Mb | RPS17 | M | 2.5 mo | 2.1/— | — | — | — | N | Short stature | Yes | |
| 802 | 3q21-tel | Variable | See¶ | M | 2 y | 7.1/21 | 0.7 | 0.72 | 103 | Y | None | Neutropenia; remission at age 16‡ | N/A |
| 802-2 | 3q21-tel | Variable | See¶ | F | 2 mo | 2.6/7.4 | 0.1 | 1.17 | 90 | N | None | Neutropenia‡ | N/A |
| 80-3§ | — | — | — | F | 2 y | 10.4/ 30.4 | 1.7 | 1 | 93 | Y | None | Neutropenia; remission at age 12‡ | N/A |
| 1786 | 15q | Variable | See# | M | 2 mo | 4.3/12.5 | — | 1.01 | — | Y | None | Remission as teenager | N/A |
ID indicates Diamond-Blackfan Anemia Registry identification number; Age, age at presentation; Hgb, hemoglobin (g/dL); Hct, hematocrit (%); Retic, reticulocyte percentage; eADA, erythrocyte adenosine deaminase activity (IU/g hemoglobin); MCV, mean corpuscular volume (fL); tel, telomere; —, data not available; ↑, reported as elevated; VSD, ventricular septal defect; and PFO, patent foramen ovale.
Regional copy variant boundaries as identified by SNP-Array/PennCNV.
The centromeric deletion boundary defined by SNP-array is conservative and does not include RPS17. The deletion boundaries identified by aCGH were 80 591 629-80 647 378 and 80 999 054-81 617 511 for sample 886 and 80 537 056-80 834 670 and 80 925 204-81 425 809 for sample 20QL. These deletion calls include both copies of RPS17 and may reflect the inability of hybridization probes to differentiate paired copies of RPS17.
All siblings have modest neutropenia (absolute neutrophil counts, 600-1200/mm3) without increased propensity for infection.
Clinical information for a second sibling is included although material for CNV analysis was not available for study.
RPL39L and RPL35A are located in the involved region on chromosome 3q; RPL35A is the only confirmed DBA gene in this region.
RPS17, RPS27L, RPS4, and RPLP1 are located on chromosome 15q; RPS17 is the only confirmed DBA gene in this region.
Single-copy deletions of RPS19 and RPS26 in DBA probands. Log2 intensity ratio of SNP probes surrounding deletions of RPS19 on chromosome 19 (A) and RPS26 on chromosome 12 (B). The regions displayed include chromosome 19 from 46 100 000-48 100 000 and chromosome 12 from 54 400 000-55 000 000, corresponding to the boxed region in the chromosome ideogram above each panel. The region of single-copy loss is shaded. The locations of RPS19 and RPS26 are indicated by the vertical line through the deletion regions.
Single-copy deletions of RPS19 and RPS26 in DBA probands. Log2 intensity ratio of SNP probes surrounding deletions of RPS19 on chromosome 19 (A) and RPS26 on chromosome 12 (B). The regions displayed include chromosome 19 from 46 100 000-48 100 000 and chromosome 12 from 54 400 000-55 000 000, corresponding to the boxed region in the chromosome ideogram above each panel. The region of single-copy loss is shaded. The locations of RPS19 and RPS26 are indicated by the vertical line through the deletion regions.
SNP-array identifies RPS17 gene deletions in DBA patients with normal r-protein gene sequence
Three patients with single-copy deletions adjacent to RPS17 were identified (Figure 2A). The centromeric deletion boundary identified by SNP-array for all 3 patients with putative RPS17 deletions was identical, beginning just telomeric to one copy of RPS17. RPS17 lies in a 200-kb intrachromosomal segmental duplication at 15q25.2, with 4 identical copies in a diploid genome.26 Limitations of SNP probe placement in the duplicated region hamper copy number estimation.
Deletions involving RPS17 in DBA probands. (A) Log2 intensity ratio of SNP probes in the deletions adjacent to RPS17 on chromosome 15. The locations of 2 identical copies of RPS17 are indicated by the vertical lines. The identified region of copy loss is adjacent to RPS17; however, the paucity of probes, notably in the HumanOmniExpress chip (20QL and 1314), is apparent. (B) Log2 intensity ratios of CGH probes in the region of RPS17. Arrows at the top of the panel indicate the positions of RPS17. Gray points and connecting line depict a 30-kb median smoothing window. The region displayed is chromosome 15 from 79 450 000-83 800 000.
Deletions involving RPS17 in DBA probands. (A) Log2 intensity ratio of SNP probes in the deletions adjacent to RPS17 on chromosome 15. The locations of 2 identical copies of RPS17 are indicated by the vertical lines. The identified region of copy loss is adjacent to RPS17; however, the paucity of probes, notably in the HumanOmniExpress chip (20QL and 1314), is apparent. (B) Log2 intensity ratios of CGH probes in the region of RPS17. Arrows at the top of the panel indicate the positions of RPS17. Gray points and connecting line depict a 30-kb median smoothing window. The region displayed is chromosome 15 from 79 450 000-83 800 000.
Given the proximity of RPS17 with the deletions identified in patients 886, 20QL, and 1314, along with the paucity of informative SNP probes in the region in question, we used aCGH to further delineate the deleted regions in 2 specimens with sufficient material for further testing (Figure 2B). Sample 20QL and 886 each showed reduced log2 probe intensity in the regions of RPS17 (Figure 2B), suggesting copy loss at one or both regions encoding RPS17. Interestingly, although limited in number, several probes in the region between the 2 copies of RPS17 appeared to have a near normal copy number. CNV detection by genomic segmentation of these aCGH studies resulted in 2 deletion calls at 15q25.2 in each patient, with each deletion call spanning a copy of RPS17. In sample 886, the called regions were 80 591 629-80 647 378 with 10 markers giving a mean marker log2 ratio of −0.72 (P = .0022) and 80 999 054-81 617 511 with 133 markers giving a mean marker log2 ratio of −0.76 (P = 9.8 × 10−45). For sample 20QL, the regions were 80 537 056-80 834 670 (15 markers; mean ratio, −0.59; P = .0013) and 80 925 204-81 425,809 (62 markers; mean ratio, −0.72; P = 3.4 × 10−18). Given the identical sequence at each copy of RPS17, probes to these exons should bind to both regions, and thus the apparent copy loss at both RPS17 regions may reflect a limitation of probe annotation (ie, probes are annotated to one region but bind at both). Therefore, although aCGH confirmed copy loss extending to RPS17, it is unclear based on these data whether 1 or 2 copies were involved in the deletion.
To further analyze the extent of RPS17 loss, we evaluated RPS17 by qPCR of both genomic DNA and mRNA in probands 20QL and 1314 (additional sample was not available from patient 886 for these studies). Compared with normal controls, copy number evaluation by qPCR showed significant reductions of RPS17 gDNA in both deletion patients that was not seen in the parents (Figure 3A). Based on the assumption of 4N normal RPS17 copy numbers, these data strongly suggest 2 copies of RPS17 for patient 20QL (mean, 0.46; SD = 0.06), however, patient 1314 could not be unambiguously assigned a 2N versus a 3N copy number based on these data (mean, 0.55; SD = 0.22). Similarly, genomic qPCR from the father of patient 1314 was consistent with either 4N or 3N copies. However, he has a normal hemoglobin concentration, MCV and eADA, as well as RPS17 mRNA expression levels that are consistent with those from unaffected individuals, suggesting that he has a normal genomic complement of RPS17 and that this finding is likely a result of assay variability. Compared with normal donors and to unaffected family members, both probands harboring RPS17 deletions showed significant reductions in RPS17 mRNA (Figure 3B).
RPS17 copy number and expression by Q-RT-PCR. (A) Copy number at RPS17 was measured after internal normalization to an intergenic region of the β-hemoglobin locus in 3 normal control individuals, the 1314 and 20QL probands, and their parents. Normal copy number (ie, 4N) was defined as the average of 3 normal control individuals in each experiment. Both DBA probands showed reduced copy number at RPS17. (B) RPS17 expression levels were measured by internal normalization to RPL35A. Both deletion probands manifest haploinsufficiency of RPS17 mRNA, with levels consistent with approximately 50% expression. Graphs depict the average of 3 independent experiments, with each control and reference gene assayed in triplicate at each experiment. Normal expression (1.0) was taken as the average of 3 normal individuals in each experiment. Error bars depict the SEM. *P < .001 and **P < .0001 by unpaired t test compared with averaged normal controls.
RPS17 copy number and expression by Q-RT-PCR. (A) Copy number at RPS17 was measured after internal normalization to an intergenic region of the β-hemoglobin locus in 3 normal control individuals, the 1314 and 20QL probands, and their parents. Normal copy number (ie, 4N) was defined as the average of 3 normal control individuals in each experiment. Both DBA probands showed reduced copy number at RPS17. (B) RPS17 expression levels were measured by internal normalization to RPL35A. Both deletion probands manifest haploinsufficiency of RPS17 mRNA, with levels consistent with approximately 50% expression. Graphs depict the average of 3 independent experiments, with each control and reference gene assayed in triplicate at each experiment. Normal expression (1.0) was taken as the average of 3 normal individuals in each experiment. Error bars depict the SEM. *P < .001 and **P < .0001 by unpaired t test compared with averaged normal controls.
Because the RPS17 gene is duplicated, resulting in 4 copies per diploid genome, it was unclear whether the loss of 1 or 2 copies of this gene would lead to an effect on ribosome synthesis. To determine whether these reductions in RPS17 copy number affected ribosome synthesis, we evaluated rRNA processing in mononuclear cells from probands 20QL and 1314. Compared with normal control and unaffected parents, the DBA probands showed steady-state increases of 21S pre-rRNA intermediate (Figure 4), which is consistent with the known effect of loss of RPS17 on pre-rRNA processing.27
Small subunit rRNA processing abnormalities in patients with RPS17 deletions. Northern blot analysis of rRNA with a hybridization probe to 18SE/ITS1 (probe γ)25 shows increased steady-state levels of 21S rRNA, indicating a functional ribosomal abnormality in patients with RPS17 deletions.
Small subunit rRNA processing abnormalities in patients with RPS17 deletions. Northern blot analysis of rRNA with a hybridization probe to 18SE/ITS1 (probe γ)25 shows increased steady-state levels of 21S rRNA, indicating a functional ribosomal abnormality in patients with RPS17 deletions.
Mosaic deletions involving DBA gene loci
In addition to segmental single-copy deletions at r-protein loci previously implicated in DBA, we identified 3 individuals from 2 families with mosaic copy loss on chromosome 3q or 15q, each with increasing levels of monosomy approaching the telomere (Figure 5). The index patient identified with a mosaic 3q abnormality is 1 of 3 siblings with DBA (Table 1). None of the patients was responsive to corticosteroids, although patient 802 and a sibling (802-3) developed spontaneous remissions in the second decade of life. A third patient (802-2) remains transfusion dependent. All siblings have modest neutropenia. Chromosomal breakage studies after treatment with mitomycin C were normal. The father had a history of unexplained anemia and sarcoidosis that were responsive to steroid treatment; he developed myelodysplastic syndrome at 44 years of age and died from complications of therapy.
Variable mosaic copy loss in DBA gene regions. BAF plots of chromosome 3 (A) and 15 (B). The heterozygous region of the BAF is displayed, showing a normal individual with most heterozygous BAFs clustering about 0.5 (top panel). A progressively increasing mosaicism is evident in patient 802 (middle panel) as a splay in the heterozygous BAF region that increases approaching the telomere. Mosaicism is more obvious in 802-2 as a bifurcation of the heterozygous BAF region (bottom panel). (B) BAF plot from a normal individual's chromosome 15q (top panel). Chromosome 15 mosaicism is evident in patient 1786 (bottom panel). The regions displayed include the long arms of the respective chromosomes, corresponding to the boxed region in the chromosome ideogram. Dashed horizontal lines in the BAF plots indicated regions grouped for mosaicism analysis. The fraction of the population with monosomy in each analyzed regions is indicated above the BAF plot in each region analyzed and is outlined in the charts to the right. The illustration on the right depicts variable chromosome 15 mosaic monosomy corresponding to the fractional mosaicism identified for sample 1786.
Variable mosaic copy loss in DBA gene regions. BAF plots of chromosome 3 (A) and 15 (B). The heterozygous region of the BAF is displayed, showing a normal individual with most heterozygous BAFs clustering about 0.5 (top panel). A progressively increasing mosaicism is evident in patient 802 (middle panel) as a splay in the heterozygous BAF region that increases approaching the telomere. Mosaicism is more obvious in 802-2 as a bifurcation of the heterozygous BAF region (bottom panel). (B) BAF plot from a normal individual's chromosome 15q (top panel). Chromosome 15 mosaicism is evident in patient 1786 (bottom panel). The regions displayed include the long arms of the respective chromosomes, corresponding to the boxed region in the chromosome ideogram. Dashed horizontal lines in the BAF plots indicated regions grouped for mosaicism analysis. The fraction of the population with monosomy in each analyzed regions is indicated above the BAF plot in each region analyzed and is outlined in the charts to the right. The illustration on the right depicts variable chromosome 15 mosaic monosomy corresponding to the fractional mosaicism identified for sample 1786.
Quantification of copy loss showed mosaic monosomy at chromosome 3q detectable in 8% of the population in the region from 125 000 000-152 000 000 and increasing 23% in the region from 187 000 000 to the telomere, the region containing RPL35A, an established DBA gene.4 Analysis of affected sibling 802-2 showed a higher proportion of mosaicism in a similar region of 3q copy loss, with monosomy from 129 000 000 to the telomere in approximately one-quarter of the peripheral blood nucleated cells (Figure 5A). A third sibling (802-3) currently in remission did not consent to testing; no 3q abnormality was detected in the mother. To assess for lineage restriction of mosaicism, we performed SNP-array analysis on lymphoid and myeloid populations from patient 802 and 802-2, which demonstrated similar levels of mosaicism in both the lymphoid and myeloid populations (data not shown).
A similar abnormality on chromosome 15q was identified in an unrelated DBA proband, patient 1786 (Figure 5B), in a region that includes RPS17. In this case, quantification of mosaicism showed < 1% monosomy of 15q in the region from 20 500 000-23 500 000 that increased to 31% of cells with monosomy over the telomeric half of the long arm. Like patient 802 with a 3q mosaic abnormality, patient 1786 developed spontaneous remission in the second decade of life.
Discussion
Defining the genetic abnormality of all DBA patients remains a critical step in understanding the molecular pathogenesis of DBA and the pathways by which the DBA phenotype is modified in different individuals, as well as for providing accurate genetic counseling to affected families and searching for less toxic treatments. The results presented here indicate that large deletions involving r-protein genes are more common in the DBA patient population than was thought previously. Given our current finding of deletions in 17% (9 of 51; 95% confidence interval, 9%-30%) of previously sequenced patients, who are estimated to account for 40% of all DBA patients, genomic deletions could be expected in 4%-10% of all DBA patients. Therefore, the frequency of genomic deletions, as a class of DBA mutation, is similar or higher in frequency then those reported for coding sequence mutations in all validated r-protein genes other than RPL5, RPS19, and RPS26.13 Furthermore, these results may underestimate the true frequency of genomic deletions. SNP-array copy number detection is limited to the extent of reliable SNP probe placement. While it is excellent in detecting large regions of CNV, loss of heterozygosity, and mosaic aneuploidy, this method may be less sensitive than some aCGH platforms for the detection of small rearrangements. Recent studies have identified smaller deletions (eg, exon-level deletions ranging from several hundred to several thousand bases) within or including r-protein genes in DBA patients.8,28 Therefore, the fraction of patients in whom known DBA r-protein genes are altered may be significantly higher than has been estimated from standard gene-sequencing studies. Our present results suggest that the inclusion of studies for the detection of genomic copy number variation at r-protein genes, SNP-array or aCGH, which are increasingly available for clinical evaluations, are an important complement to sequencing studies in the molecular diagnosis of DBA. Furthermore, although a considerable majority of cases of DBA are now attributable to abnormalities in genes encoding r-proteins, our results suggest that abnormalities in non-r-protein genes should still be considered as candidate genes for DBA.
Copy number variations at 1 large and 3 small subunit r-protein genes with established importance in DBA was identified in this study: RPL35A, RPS19, RPS26, and RPS17. RPS19 and RPS26 are commonly mutated in DBA, with frequencies estimated at 25% and 10%, respectively,7,29 whereas mutations of RPS17 are far less common, with only 4 mutations reported in 262 patients (1.5%).3,5,30,31 The identification of 3 similar RPS17 deletions in unrelated individuals in this study, along with recent reports of 5 additional RPS17 deletions,8,28 is higher (5.5%) than anticipated from mutational analysis and suggests this region may be vulnerable to copy variation. Furthermore, copy loss involving RPS17, along with mental retardation and hypoproliferative anemia suggestive of DBA, was reported in one patient in a study evaluating CNVs in patients with mental retardation.32
Both genomic copy gains and losses involving RPS17 are observed in patients without a hematologic phenotype and in normal controls, suggesting the possibility of individual variability in sensitivity to copy change around RPS17.24,26,33-35 A review of Database of Genomic Variants data, outlined in Table 2, demonstrates copy loss at several additional r-protein gene regions, including genes that have been potentially implicated in DBA. These data, from putatively normal controls, should be interpreted with caution, since DBA is well-known to have variable penetrance, with some unaffected or minimally affected family members sharing a pathogenic mutation with classically affected relatives. Furthermore, tolerance of copy loss does not exclude the possibility that mutations in these genes may act through a dominant-negative mechanism rather than through allelic haploinsufficiency (eg, the missense RPS15 mutation).5 Nonetheless, the high frequency of copy loss around genes such as RPL3L or RPS11 suggests that these are unlikely to be DBA genes. In the case of RPS17, reports of missense and nonsense mutations in DBA probands from multiple populations, along with the relatively high frequency of copy loss outlined above, suggest a pathogenic effect. However, this interpretation is further confounded by the observation of 4 apparently functional copies of RPS17.
Copy loss at r-protein and r-protein-like genes reported in the Database of Genomic Variants
| Gene . | No with copy loss . | No studied . | % with loss . | Studies reported . |
|---|---|---|---|---|
| RPLP2 | 16 | 580 | 2.7 | 2 |
| RPL3L | 29 | 95 | 30.5 | 1 |
| RPL146 | 1 | 112 | 0.8 | 1 |
| RPL196 | 10 | 95 | 10.5 | 1 |
| RPL22 | 3 | 95 | 3.2 | 1 |
| RPL31 | 13 | 2906 | 0.4 | 1 |
| RPL36AL | 4 | 450 | 0.9 | 1 |
| RPS4X | 1 | 90 | 1.1 | 1 |
| RPS5 | 24 | 2573 | 0.9 | 4 |
| RPS9 | 30 | 1604 | 1.9 | 3 |
| RPS11 | 47 | 220 | 21.4 | 3 |
| RPS155 | 42 | 175 | 24 | 3 |
| RPS173,5,30,31 | 6 | 405 | 1.5 | 4 |
| RPS21 | 20 | 3605 | 0.6 | 4 |
| Gene . | No with copy loss . | No studied . | % with loss . | Studies reported . |
|---|---|---|---|---|
| RPLP2 | 16 | 580 | 2.7 | 2 |
| RPL3L | 29 | 95 | 30.5 | 1 |
| RPL146 | 1 | 112 | 0.8 | 1 |
| RPL196 | 10 | 95 | 10.5 | 1 |
| RPL22 | 3 | 95 | 3.2 | 1 |
| RPL31 | 13 | 2906 | 0.4 | 1 |
| RPL36AL | 4 | 450 | 0.9 | 1 |
| RPS4X | 1 | 90 | 1.1 | 1 |
| RPS5 | 24 | 2573 | 0.9 | 4 |
| RPS9 | 30 | 1604 | 1.9 | 3 |
| RPS11 | 47 | 220 | 21.4 | 3 |
| RPS155 | 42 | 175 | 24 | 3 |
| RPS173,5,30,31 | 6 | 405 | 1.5 | 4 |
| RPS21 | 20 | 3605 | 0.6 | 4 |
Data from Database of Genetic Variants Version 10 are available at: http://projects.tcag.ca/variation.24 Citations indicate genes with reported associations with DBA. Database entries where 1 sample of 1 tested showed copy loss and entries reporting copy gain are not included.
The majority of r-protein genes exist in the mammalian genome as single-copy genes, typically with numerous processed pseudogenes. Several r-protein genes are duplicated, RPL10 and RPL10L, RPL26 and RPL26L, RPL36, and RPL36L, for example, with similar genomic sequence encoding paralogous proteins, the functions of which are largely unknown.36 To date, no pathogenic mutations have been conclusively established in these duplicated genes. In the case of RPS17, the 2 copies are precisely duplicated with 100% sequence identity over the 3.7-kb genomic structure and > 99% identity over the 200-kb segmental duplication. Therefore, RPS17 sequencing studies should detect both copies. Although previous RPS17 sequencing studies report heterozygous mutations, none provided sufficient data to quantify at the allele level whether 1, 2, or 3 copies of RPS17 are abnormal. These studies have also not addressed the potential functional consequences of these mutations on ribosome synthesis. Therefore, despite having clearly identifiable loss-of-function mutations in at least 1 copy of RPS17, the presence of 4 copies per diploid genome raises critical questions concerning the potential role of these mutations in DBA pathogenesis. Are all 4 copies of RPS17 transcriptionally active? Could patients with apparently heterozygous RPS17 mutations also harbor RPS17 deletions? Might identical RPS17 mutations occur at different copies of RPS17, suggesting a gene conversion event?
To address some of these issues, we sought to clarify RPS17 gene dosage, expression level, and small subunit rRNA processing patterns in 2 patients with putative RPS17 deletions. Evaluation of gDNA from patient 20QL showed precisely half-normal values of RPS17 gDNA, strongly suggesting 2 residual copies of RPS17 (Figure 3A). Similarly, evaluation of RPS17 expression showed an approximately 50% reduction in RPS17 mRNA levels (Figure 3B). Evaluation of pre-rRNA processing in patient 20QL revealed an increased ratio of 21S to 18SE pre-rRNA relative to parents and normal controls (Figure 4), a pattern predicted by RPS17 knock-down studies in HeLa cells.27,37 Therefore, the reduction from 4 to 2 copies of RPS17 in patient 20QL is clearly associated with a functional disruption of ribosome synthesis. Evaluation of patient 1314 also showed reduced RPS17 copy number and expression at a level consistent with either 2 or 3 copies; the experimental variance in this sample was higher, the differences between 1314 and normal controls were less statistically significant than for 20QL, and the absolute values for expression and copy number were higher in sample 1314 than in 20QL. Analysis of pre-rRNA processing for sample 1314 showed increased accumulation of 21S pre-rRNA relative to controls, although this was less dramatic than that observed in patient 20QL. One potential explanation of these data could be that patient 1314 retains 3 copies of RPS17. However, it is important to note that there is no obvious phenotypic characteristic distinguishing these patients. There are numerous examples in the DBA literature of seemingly inexplicable individual variations in the face of similar or identical underlying genetic lesions. We anticipate that with the identification of additional patients with RPS17 deletions and mutations, the question of RPS17 dose in DBA can be more thoroughly explored.
The identification of chromosome-specific variable mosaicism was an unanticipated finding and is a novel observation in DBA. An advantage of SNP-array over other methods of genomic copy number analysis is the ability to detect mosaic aneuploidy as well as copy-neutral loss of heterozygosity. In a normal dizygous chromosome, the BAF plot shows data clustered around the 1, 0.5, and 0 frequencies, corresponding to the BB, AB, and AA genotypes. With mosaic monosomy, the heterozygous B-allele frequency splits symmetrically at approximately the 50% region, with SNPs at which an A allele is lost shifting toward 1 and those at which the B allele is lost shifting toward 0 and with the degree of shift proportional to the fraction of mosaicism.38 Whereas whole-chromosome mosaic aneuploidy has been detected using SNP-array, the cases reported here differ in that the proportion of cells showing mosaic copy loss varies, with increases approaching the telomere. These findings suggest a chromosome-specific instability leading to localized, progressive loss of genomic material.
Several aspects of these observations suggest that this unusual form of chromosomal abnormality is directly related to DBA in these patients: (1) the identification of a similar structural abnormality in 2 unrelated DBA families, (2) the presence of known DBA r-protein genes in the regions of mosaic copy loss, and (3) the finding of a similar region of genomic copy loss in 2 related individuals with DBA. Also noteworthy is the fact that 2 probands with variable mosaicism experienced a spontaneous remission of DBA, whereas sibling 802-2, who currently remains transfusion dependent, has a larger region and a higher fraction of mosaicism. The pedigree and finding of 2 similar abnormalities in affected members in the 3q mosaic deletion kindred strongly support an autosomal-dominant mode of inheritance. The demonstration of comparable myeloid and lymphoid mosaicism further supports an inherited defect, in contrast to a recently identified somatic mosaic loss of RPS14 in the 5q syndrome, where an acquired 5q abnormality was demonstrated by copy loss limited to the circulating myeloid fraction.39 The 15q mosaic deletion case was sporadic, with no family history of DBA or unexplained anemia. SNP-array analysis in this case was limited to unfractionated circulating DNA. It is unclear, based on presently available data, whether the mosaic abnormality at 15q represents an inherited or acquired form of DBA.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by grants from the National Institutes of Health (K08HL092224 to J.E.F., R01HL079571 to A.V. and J.M.L.); by the National Heart, Lung, and Blood Institute DNA Resequencing and Genotyping Service (R109 MOHLKE to A.V., J.M.L., J.E.F., R.J.A., and S.R.E.); by the Feinstein Institute for Medical Research General Clinical Research Center (M01RR018535 to A.V. and J.M.L.); and by National Human Genome Research Institute intramural funds (to D.M.B.).
National Institutes of Health
Authorship
Contribution: J.E.F. developed the project, designed and performed the research, analyzed the data, and drafted the manuscript; A.V., E.A., and J.M.L. developed the project, designed the research, collected and analyzed the clinical data, and edited the manuscript; H.C.-D. and T.C.M. contributed analytic methodology and analyzed the data; R.J.A. analyzed the data and edited the manuscript; S.R.E. designed and performed the research, analyzed the data, and edited the manuscript; and D.M.B. developed and supervised the project, analyzed the data, and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jason Farrar, MD, 1650 Orleans St, CRB I Rm 209, Baltimore, MD 21231; e-mail: jfarrar4@jhmi.edu; or David Bodine, PhD, Bldg 49, Rm 4A04, 49 Convent Dr, MSC 4442, Bethesda, MD 20892; e-mail: tedyaz@mail.nih.gov.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal