Abstract
Although anemia is common in Shwachman- Diamond syndrome (SDS), the underlying mechanism remains unclear. We asked whether SBDS, which is mutated in most SDS patients, is critical for erythroid development. We found that SBDS expression is high early during erythroid differentiation. Inhibition of SBDS in CD34+ hematopoietic stem cells and early progenitors (HSC/Ps) and K562 cells led to slow cell expansion during erythroid differentiation. Induction of erythroid differentiation resulted in markedly accelerated apoptosis in the knockdown cells; however, proliferation was only mildly reduced. The percentage of cells entering differentiation was not reduced. Differentiation also increased the oxidative stress in SBDS-knockdown K562 cells, and antioxidants enhanced the expansion capability of differentiating SBDS-knockdown K562 cells and colony production of SDS patient HSC/Ps. Erythroid differentiation also resulted in reduction of all ribosomal subunits and global translation. Furthermore, stimulation of global translation with leucine improved the erythroid cell expansion of SBDS-knockdown cells and colony production of SDS patient HSC/Ps. Leucine did not reduce the oxidative stress in SBDS-deficient K562 cells. These results demonstrate that SBDS is critical for normal erythropoiesis. Erythropoietic failure caused by SBDS deficiency is at least in part related to elevated ROS levels and translation insufficiency because antioxidants and leucine improved cell expansion.
Introduction
Shwachman-Diamond syndrome (SDS) is a rare multisystem disorder, primarily affecting the bone marrow, pancreas, and skeletal systems.1,2 Hematologic abnormalities are a major cause for morbidity and mortality and include cytopenia, myelodysplastic syndromes, and leukemia. Neutropenia is observed in almost all SDS patients. Approximately 85% of the patients have hypomorphic mutations in the Shwachman Bodian Diamond syndrome gene, SBDS.3,4 Knockdown of the SBDS homolog in murine myeloid 32Dcl3 cells resulted in normal neutrophil maturation but reduced viability of granulocyte precursors.5 These studies provide evidence that SBDS is critical for normal granulopoiesis. The role of SBDS in erythroid development has not been characterized. Anemia is observed in ∼ 60% of SDS patients. In addition, 60% of the patients have high erythrocyte mean corpuscular volume, and 70% have high fetal hemoglobin blood levels.4,6,7 Despite these erythrocyte abnormalities, SDS has frequently been classified as an inherited neutropenia disorder.8,9 The underlying mechanism of anemia and the degree to which it can be attributed to erythropoietic failure, nutritional deficiencies, and/or recurrent infection have not been studied.
Most SDS bone marrows are hypocellular with reduced hematopoietic progenitors, including erythroid cells. Nevertheless, the defects mediated by SBDS deficiency that are responsible for this phenotype are unclear. For example, it is unknown whether the reduction in SDS hematopoietic progenitors is the result of an impaired ability to differentiate or to proliferate or because of a defect to survive throughout the process of maturation. Previous studies support SBDS roles in several cellular pathways, including cell survival.10,11 Studies on SDS marrow cells similarly showed reduced hematopoietic stem cells/early progenitors (HSC/Ps) and colony formation.12 In both SDS marrow cells and SBDS-knockdown HeLa cells, the slow cell expansion was in part the result of Fas-mediated apoptosis.13 Importantly, inhibition of apoptosis in HeLa cells rescued the slow cell expansion phenotype. Elevated levels of reactive oxygen species (ROS) may affect erythroid cell survival and can cause cytopenia.14,15 We have previously found that knockdown of SBDS in myeloid and nonmyeloid cell lines causes elevated ROS levels, and balancing ROS levels improved cell survival and expansion.16 Although these studies shed light on the role of SBDS in cell survival, they were not performed on differentiating cells and may not accurately represent the SBDS role during maturation of hematopoietic cells.
A role of SBDS in promoting ribosome biogenesis has also been suggested.17-20 The yeast homolog has been shown to play a role in maturation of the 60S ribosome subunit.18 However, the dynamics of ribosome biogenesis during hematopoiesis and how these defects relate to the hematologic abnormalities seen in SDS are unknown. Because the developing erythroid cells are highly proliferative and require increased ribosome synthesis for the increased demand of hemoglobin synthesis, it is possible that these immature cells are more dependent on SBDS than mature cells.
In this study, we asked whether, in addition to its role in granulocytic cell development, SBDS is also critical for the expansion and differentiation of developing erythroid progenitors. Because expansion capability was reduced, we studied whether apoptosis and proliferation of differentiating erythroid cells are affected when SBDS is depleted. We found that apoptosis was predominantly affected and was accelerated. We then asked whether the cell expansion phenotype is dependent on defects in global translation and ROS levels. In this study, we focused on the erythroid lineage model, to ask whether the anemia in SBDS-deficient cells is the result of hematopoietic failure and to perform an in-depth analysis of the underlying defects leading to cytopenia in SDS.
Methods
Subjects
All SDS patients had characteristic hematologic and pancreatic manifestations,7 biallelic SBDS mutations, and no signs of leukemic transformation or clonal marrow cytogenetic abnormalities. Healthy control subjects were donors for bone marrow transplantation. Cord blood was obtained from women after labor. Informed written consent was obtained from all patients and controls or their legal guardians in accordance with the Declaration of Helsinki. The studies were approved by the Hospital for Sick Children, University of Toronto Research Ethics Board.
Cell culture and erythroid differentiation
CD34+ HSC/Ps from marrow mononuclear cells were enriched using immunomagnetic beads.11,12 CD133+ HSC/Ps from cord blood samples were sorted using an anti-CD133/2–PE antibody (Miltenyi Biotec). HSC/Ps were plated in serum-free granulocyte-erythrocyte-monocyte-megakaryocyte colony-forming unit assays.11 In other experiments, HSC/Ps were plated in differentiation cultures containing IMDM, 20% FBS, 2 U/mL erythropoietin, 50 ng/mL SCF, and 20 μU/mL insulin, followed by addition of erythropoietin every 2 days.
K562 cells were from ATCC. To induce erythroid differentiation, K562 cells were seeded at a density of 2 × 105 cells/mL in IMDM (Invitrogen) with FBS and 25 μm hemin solution (Sigma-Aldrich).21
shRNA expression cassettes and generation of SBDS-knockdown cells
SBDS short hairpin RNA (shRNA) and a scrambled (SCR) RNA control sequence were cloned into a pSEC/Neo plasmid (Ambion) as previously described.10 K562 cells were transfected with the plasmids and selected by plating in methylcellose Cell Selection medium (StemCell Technologies) containing geneticin (Invitrogen), and individual colonies were isolated after 14 days. Two knockdown lines (K562/shSBDS-1 and K562/shSBDS-3) and 1 control line (K562/shSCR) were established.
Viral vectors production and transduction
We cloned SBDS in frame with YPet on a pLNCX/YPet vector (courtesy of Dr Patrick Daugherty, University of California, Santa Barbara, CA). Viral particles were produced by transfecting FLYRD18 retroviral packaging cells22 with pLNCX retroviral vectors expressing SBDS-YPET and control YPET. K562/shSBDS-3 cells were transduced in the presence of polybrene. K562/shSBDS-3 cells express an shRNA targeting nucleotides 137 to 156 3′ to the open reading frame; therefore, the reintroduced SBDS open reading frame was not inhibited by the shRNA.
The shRNA oligonucleotides, which were used to target SBDS in K562 cells, were also cloned into the YFP vector, pFCYsi (courtesy of Dr Dan Link, Washington University). To knock down SBDS in human HSC/Ps, marrow CD34+ cells from healthy control subjects were incubated in StemSpan medium (StemCell Technologies) with FMS-like tyrosine kinase 3 ligand, SCF, and thrombopoietin for 24 hours. Cells were then transduced 3 times with viral supernatant in the presence of polybrene.23
Morphologic analysis by benzidine and Wright-Giemsa staining
Benzidine staining was used to detect pseudo-peroxidase activity of hemoglobin.21 Cells were cytospun on glass slides and incubated with 3,3′-dimethoxybenzidine (Sigma-Aldrich) and hydrogen peroxide. Benzidine-positive cells were enumerated by light microscopy. Cytologic examination of bone marrow was done after Wright-Giemsa staining.24 The proportion of the various marrow cell populations was determined by counting at least 500 cells under light microscope.
Cell expansion assays
K562 cells were seeded at 2.0 × 104 cells per 0.1 mL in 96-well plates or 2.0 × 105 cells/mL in 35-mm dishes depending on the specific experiment. At the indicated time points after plating, viable cell counts were determined using either trypan blue dye exclusion or MTT assay (ATCC).11 Absorbance was measured at 570 nm in a microplate reader (Bio-Tek Instruments).
RNA quantitative PCR
Expression of SBDS, GATA-1, and TFR2 was measured by RNA quantitative real-time PCR using SYBR Green technology.25 The specific primer pairs for the studied genes and the internal control ACTB gene are described in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). In each experiment, no-DNA template control was also used. Relative expression levels were calculated using the relative standard curve method.26
Western blotting
Western blot analysis was performed as previously described.11 Chicken anti–human SBDS antibody was generated as previously described,10 Mouse anti–human β-actin antibody was from Sigma-Aldrich; mouse anti–human p53 antibody was from Santa Cruz Biotechnology; mouse anti-p63 (A4A) was from Santa Cruz Biotechnology; mouse anti-TAp73 (BL-906) was from Bethyl Laboratories; a rabbit anti–human cleaved poly(ADP-ribose) polymerase (Asp214) antibody was from Cell Signaling Technology.
Flow cytometry
Propidium iodide and Ki-67 costaining was performed as previously described10 with minor modifications. In brief, 5 × 105 cells were washed with PBS and fixed with 70% ethanol. Cells were rewashed and incubated with mouse anti–Ki-67 (Dako Canada) at 4°C for 30 minutes in the dark, followed by incubation with FITC-conjugated IgM antibody (GE Healthcare) for 30 minutes. Cells were rewashed and incubated 1 hour with propidium iodide, RNaseA, and Triton X-100. DNA content was analyzed by flow cytometry, and the sub-G1 cell populations were quantified, whereas Ki-67-FITC expression was analyzed in viable cells.
For evaluation of intracellular ROS level, cells (5 × 105) were washed and incubated for 30 minutes with 50M 2,7-dichlorodihydrofluorescein diacetate (Cayman Chemical). Cells were washed with HBSS and analyzed by flow cytometry within 15 minutes.
Ribosome profile analysis by sucrose gradient density ultracentrifugation
Cells were incubated at 37°C for 10 minutes after treating with cycloheximide, washed in cold PBS containing cycloheximide, and resuspended in TMK100 lysis buffer (Tris, MgCl2, KCl, HEPES, cycloheximide, Triton X-100, and RNAseOUT).27 Nuclei were removed by centrifugation at 12 000g for 5 minutes. Lysates were layered on a 5% to 45% (weight/volume) sucrose gradient in sucrose gradient buffer containing KCl, MgCl2, and HEPES and centrifuged at 197 000g for 2.5 hours. To evaluate the ratio of 40S to 60S, cells were lysed with EDTA instead of MgCl2 to dissociate polysomes and mRNA. Gradients were centrifuged for 3 hours at 197 000g and fractionated using the Brandel gradient fractionator system (Brandel). Absorbance was monitored at 254 nm with an ISCO UA-6 flow cell (Teledyne ISCO).
Evaluation of global translation by incorporation of 35S methionine/cysteine
Metabolic labeling of K562 cells with [35S]-methionine was performed as previously published27 with minor modifications. Cells were incubated with [35S]methionine and placed for 30 minutes at 37°C. The labeling medium was removed, and the cells were washed and lysed. [35S]methionine incorporation was determined by trichloroacetic acid precipitation of the supernatant aliquots. Counts per minute were obtained using a scintillation counter (Beckman Coulter LS 6500).
Oligonucleotide microarray
Ficoll-extracted marrow mononuclear cells from 9 SDS patients and 7 healthy subjects were analyzed by oligonucleotide microarray using the HG_U133_Plus 2.0 GeneChip (Affymetrix).17 Data analysis and generation of moderated T-statistics value were performed as previously described.17 Genes with an average probe T-statistics value of either higher than 1.7 or lower than −1.7 were considered candidate ROS-regulating genes in SDS. The average percentages of the various bone marrow cell lineages: myeloid, erythroid, lymphoid, and other cells were in the normal range (± SD): 46.4% ± 6.8%, 31.1% ± 7.3%, 19.5% ± 5.5%, and 3.0% ± 1.3%, respectively. Differential expression of genes involved in leukemia, apoptosis, and ribosome biogenesis were reported elsewhere.10,17,25 All microarray data are available on the Gene Expression Omnibus under accession number GSE32057.
Statistics
Student t test was used to determine the statistical significance of differences between 2 means. A 1-way ANOVA followed by Dunnett test or Tukey test of multiple means was performed to determine statistical significance of differences between multiple means.
Results
SBDS is expressed early during erythroid differentiation
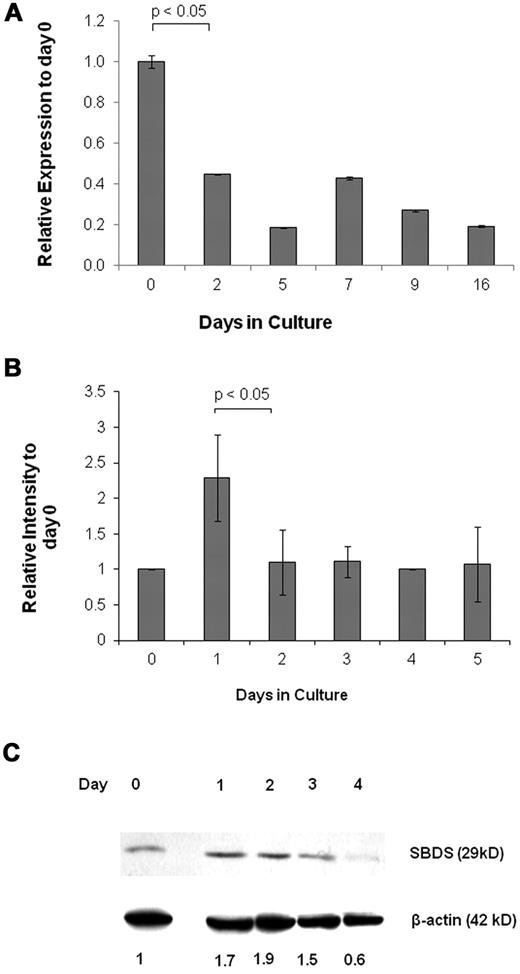
To determine whether SBDS is expressed at a particular stage of erythroid development, we analyzed its expression by quantitative RNA-PCR. Early CD133+/HSC/Ps showed high expression of SBDS, followed by a decline in expression as erythroid differentiation progressed (Figure 1A). We also used K562 human erythroleukemia cells as a model for erythroid development because they are multipotent hematopoietic progenitors, which can differentiate to normoblasts over ∼ 5 days using agents, such as hemin.28 SBDS was up-regulated 24 hours after induction of differentiation. Expression returned to basal levels for the subsequent 4 days (Figure 1B). Increased SBDS protein levels in K562 were detected one day after the mRNA levels peaked (Figure 1C). Morphologic examination of undifferentiated and differentiating cells is shown in supplemental Figure 1A and B.
SBDS expression during erythroid differentiation. (A) Real-time RNA quantitative PCR was used to analyze daily SBDS mRNA expression in CD133+ HSC/Ps after induction of erythroid differentiation. The experiment was performed in triplicate. The ACTB gene was used as an internal control. (B) SBDS mRNA expression by RNA quantitative PCR was also determined in wild-type K562 cells after induction of erythroid differentiation by hemin. Three separate experiments were conducted in triplicate. (C) SBDS protein expression in hemin-induced K562 cells by Western blotting (representative blot of 3 experiments).
SBDS expression during erythroid differentiation. (A) Real-time RNA quantitative PCR was used to analyze daily SBDS mRNA expression in CD133+ HSC/Ps after induction of erythroid differentiation. The experiment was performed in triplicate. The ACTB gene was used as an internal control. (B) SBDS mRNA expression by RNA quantitative PCR was also determined in wild-type K562 cells after induction of erythroid differentiation by hemin. Three separate experiments were conducted in triplicate. (C) SBDS protein expression in hemin-induced K562 cells by Western blotting (representative blot of 3 experiments).
SBDS deficiency results in markedly reduced expansion capability of differentiating erythroid cells
We studied the effect of SBDS deficiency on expansion of primary human erythroid cells by plating SBDS-knockdown marrow CD34+ cells in erythroid differentiation medium. SBDS-knockdown erythroid cells showed impaired expansion compared with controls (Figure 2A). Effective protein depletion is shown in supplemental Figure 2A. Because these experiments apply transient transduction of primary cells with limited life span, the viability and expansion of the scrambled RNA control are slightly compromised compared with nontransduced wild-type cells. We also studied BFU-E colony growth in clonogenic assays. CD34+ cells from SDS patients produced less BFU-E colonies than normal controls (SDS, median ± SEM = 7 ± 3 colonies/103 CD34+ cells, n = 3; and normal, 17 ± 4 colonies, n = 5, P = .04).
Cell expansion of SBDS-deficient cells during erythroid differentiation. (A) SBDS-knockdown and control CD34+ HSC/Ps were induced to undergo erythroid differentiation by a combination of SCF and erythropoietin and cell counts were monitored daily by trypan blue exclusion. (B) SBDS-knockdown K562/shSBDS-3 cells were transduced with a pLNCX retrovirus expressing YPet and SBDS open reading frame (which is not targeted by the shRNA expressed). YPet-positive cells were sorted and plated at a concentration of 2 × 105 cells/mL and treated with 25 μm hemin for 5 days. Cell expansion was assessed by trypan blue exclusion. Data are the mean ± SE of 3 independent experiments. *P < .05, statistically significant results using 1-way ANOVA and Tukey test for multiple means against both S-3 + YPET and S-3 cells.
Cell expansion of SBDS-deficient cells during erythroid differentiation. (A) SBDS-knockdown and control CD34+ HSC/Ps were induced to undergo erythroid differentiation by a combination of SCF and erythropoietin and cell counts were monitored daily by trypan blue exclusion. (B) SBDS-knockdown K562/shSBDS-3 cells were transduced with a pLNCX retrovirus expressing YPet and SBDS open reading frame (which is not targeted by the shRNA expressed). YPet-positive cells were sorted and plated at a concentration of 2 × 105 cells/mL and treated with 25 μm hemin for 5 days. Cell expansion was assessed by trypan blue exclusion. Data are the mean ± SE of 3 independent experiments. *P < .05, statistically significant results using 1-way ANOVA and Tukey test for multiple means against both S-3 + YPET and S-3 cells.
Similarly, we studied the expansion of SBDS-knockdown K562 cells (supplemental Figure 2B). Western blotting confirmed approximately 5% residual protein in K562/shSBDS-3 cells and approximately 12% in K562/shSBDS-1 cells (supplemental Figure 2C). After induction of erythroid differentiation, the expansion of the SBDS-knockdown cells was markedly reduced compared with control cells (P < .01 for K562/shSBDS-1 and P < .001 for K562/shSBDS-3). To determine whether the reduced cell expansion phenotype was a direct consequence of SBDS deficiency, SBDS (that is not targeted by the shRNA expressed in K562/shSBDS-3) was reintroduced into K562/shSBDS-3 cells. SBDS expression completely rescued cell expansion (Figure 2C). The higher numbers of cells after reintroduction of SBDS to the knockdown cells on day 3 and 4 might be related to higher SBDS expression levels during these days. Transduction efficiency is shown in supplemental Figure 2D and E.
SBDS-deficient cells retain the ability to enter into a differentiation program
Because reduction of SDS marrow cells can be the result of early arrest in differentiation, we asked whether SBDS deficiency impairs the entry into an erythroid differentiation program. First, we analyzed the relative percentage of different hematopoietic cells in marrows from SDS patients (Table 1). Although the patients had reduced erythropoietic cells because of the hypoplastic bone marrows, the percentages of erythroid marrow population and normoblast subpopulation (basophilic, polychromatic, and orthocromatic) were in the normal range. The percentage of pro-erythroblasts in most patients was mildly elevated. Next, we examined the ability of SBDS-deficient K562 cells to undergo hemoglobinization after induction of differentiation. The percentages of hemoglobinized (benzidine-positive) SBDS-knockdown cells were higher than the control cells (supplemental Figure 3A-B) with a significant difference between K562/shSBDS-S3 and K562/shSCR cells on day 5. The expression of the transcription factors GATA1 and TFR2, which are critical for erythroid development in SBDS-knockdown cells, was also increased in the SBDS-knockdown cells (supplemental Figure 3C).
Bone marrow differential of patients with SDS
| Patient (mutation)* . | Age, y . | Proerythroblasts, % . | Normoblasts (basophilic, polychromatic, orthochromatic), % . | Promyelocytes, myelocytes, metamyelocytes, %) . | Bands, neutrophils, % . | Lymphocytes, % . | Myeloblasts, % . | Cellularity,† % . | Peripheral blood Hg, g/L; reticulocytes, × 109/L; neutrophils, × 109/L . |
|---|---|---|---|---|---|---|---|---|---|
| 1 (A/A + B) | 21.5 | 5 | 28 | 29 | 6 | 6 | 2.5 | 33-66 | 72;28;1.1 |
| 2 (A/A + B) | 13.5 | 7 | 31.5 | 37.5 | 6 | 15.5 | 2.5 | 33-66 | 90;ND;1.05 |
| 3 (A/B) | 20 | 2 | 21.5 | 44.5 | 7.5 | 23 | 2.5 | Indeterminate | 91;ND;2.4 |
| 4 (A/C) | 20 | 2 | 24 | 40 | 5 | 28 | 1 | 33-66 | 123;ND;1.2 |
| 5 (A/D) | 22.5 | 7.5 | 30 | 31.5 | 10 | 17.5 | 2.5 | < 33 | 114;44;0.56 |
| 6 (A/A + B) | 4 | 5 | 21 | 33.5 | 18.5 | 18 | 2.5 | > 66 | 113;1;0.47 |
| 7 (A/B) | 13.5 | 1 | 23 | 40 | 21 | 14 | 2 | Indeterminate | 164;4;3.0 |
| 8 (A/B) | 31.5 | 2 | 39 | 31 | 15.5 | 15 | 2 | 33-66 | 120;ND;0.9 |
| Normal54 | > 12 | 0.2-1.3 | 18.8-36.2 | 19.6-34.2 | 13.5-27.3 | 11.1-23.2 | 0.2-1.5 |
| Patient (mutation)* . | Age, y . | Proerythroblasts, % . | Normoblasts (basophilic, polychromatic, orthochromatic), % . | Promyelocytes, myelocytes, metamyelocytes, %) . | Bands, neutrophils, % . | Lymphocytes, % . | Myeloblasts, % . | Cellularity,† % . | Peripheral blood Hg, g/L; reticulocytes, × 109/L; neutrophils, × 109/L . |
|---|---|---|---|---|---|---|---|---|---|
| 1 (A/A + B) | 21.5 | 5 | 28 | 29 | 6 | 6 | 2.5 | 33-66 | 72;28;1.1 |
| 2 (A/A + B) | 13.5 | 7 | 31.5 | 37.5 | 6 | 15.5 | 2.5 | 33-66 | 90;ND;1.05 |
| 3 (A/B) | 20 | 2 | 21.5 | 44.5 | 7.5 | 23 | 2.5 | Indeterminate | 91;ND;2.4 |
| 4 (A/C) | 20 | 2 | 24 | 40 | 5 | 28 | 1 | 33-66 | 123;ND;1.2 |
| 5 (A/D) | 22.5 | 7.5 | 30 | 31.5 | 10 | 17.5 | 2.5 | < 33 | 114;44;0.56 |
| 6 (A/A + B) | 4 | 5 | 21 | 33.5 | 18.5 | 18 | 2.5 | > 66 | 113;1;0.47 |
| 7 (A/B) | 13.5 | 1 | 23 | 40 | 21 | 14 | 2 | Indeterminate | 164;4;3.0 |
| 8 (A/B) | 31.5 | 2 | 39 | 31 | 15.5 | 15 | 2 | 33-66 | 120;ND;0.9 |
| Normal54 | > 12 | 0.2-1.3 | 18.8-36.2 | 19.6-34.2 | 13.5-27.3 | 11.1-23.2 | 0.2-1.5 |
A indicates 258 + 2T > C; B, 183-184TA > CT; C, 260T > G; and D, 203A > G.
The cellularity was determined as the percentage of areas with hematopoietic cells on bone marrow biopsies. In cases where no biopsy was available, the percentage of areas of hematopoietic cells on aspirate fragments was determined. In case of no fragments seen on the aspirates, the cellularity was deemed “indeterminate.”
Ablation of SBDS results in markedly increased apoptosis during erythroid differentiation
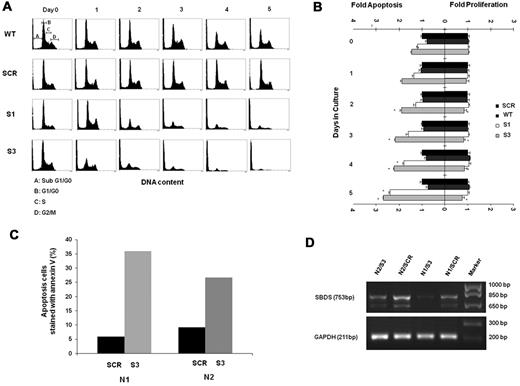
The underlying mechanisms responsible for reduced cell expansion can be either accelerated cell death and/or reduced proliferation. We first studied the viability of differentiating SBDS-knockdown by the MTT assay. The MTT yellow tetrazole dye is reduced to purple formazan in living cells. We found a dramatic reduction of viability in the SBDS-knockdown cells compared with controls during differentiation (supplemental Figure 4A). Next, sub-G0/G1 apoptotic cells were determined using a propidium iodide binding assay.10,29 Apoptosis was slightly increased in the nondifferentiating SBDS-knockdown cells (Figure 3A-B). For example, the percentage of apoptotic K562/shSBDS-3 cells was 1.46-fold higher than that of K562/shSCR cells (P < .05). After induction of differentiation, apoptosis was markedly increased in both knockdown cell lines. For example, the percentage of apoptotic K562/shSBDS-3 cells was 2.68-fold higher than that of K562/shSCR cells (P < .01). Caspase-3 activation as determined by semiquantitative Western blotting of cleaved poly(ADP-ribose) polymerase was also enhanced in SBDS-knockdown K562 cells compared with controls in both undifferentiated and differentiated cells (supplemental Figure 4B). In addition, we transduced CD34+ cells with either an shSBDS-3 or shSCR cassette, induced erythroid differentiation, and evaluated apoptosis by annexin V staining 5 days after plating. CD34+ cells from 2 different healthy persons manifested 6- and 3-fold higher rates of apoptosis when shSBDS was transduced compared with transduction with shSCR (Figure 3C-D).
Apoptosis and proliferation levels in differentiating SBDS-deficient cells. (A) Control and knockdown K562 cells were induced to undergo erythroid differentiation for 5 days. Cells were fixed in 70% ethanol and stained with propidium iodide for DNA content analysis. A representative diagram of 4 independent experiments is shown. Pre-G0/G1 cells represent apoptotic cells. Representative distributions of the cell cycle phases are depicted on day 0 of wild-type cells. (B) Cells were costained for propidium iodide to evaluate pre-G0/G1 apoptotic cells and for Ki-67 expression on nonapoptotic cells to evaluate proliferating cells. Cells were then analyzed by flow cytometry. Comparison of the mean fold increase in apoptosis versus mean fold decrease in proliferation of 4 independent experiments is shown. The values for apoptosis and proliferation were normalized to the value obtained for each day in control K562 cells, and assigned a value of 1.0 for proliferating cells and apoptotic cells. (C) Mobilized CD34+ cells from 2 donors for hematopoietic stem cell transplantation were purified, transduced, and induced to undergo erythroid differentiation as described in “Cell culture and erythroid differentiation.” After 5 days, cells were harvested and evaluated for apoptosis rates using annexin V staining. For each donor cell (N1 and N2), annexin V staining of cells transduced with scrambled RNA controls (SCR) or shRNA against SBDS (S-3) are shown. (D) Results of inhibition of SBDS mRNA in CD34+ cell samples are shown. The faster running SBDS band corresponds to a previously published shorter isoform. (Cell numbers did not allow corresponding testing of SBDS protein levels.)
Apoptosis and proliferation levels in differentiating SBDS-deficient cells. (A) Control and knockdown K562 cells were induced to undergo erythroid differentiation for 5 days. Cells were fixed in 70% ethanol and stained with propidium iodide for DNA content analysis. A representative diagram of 4 independent experiments is shown. Pre-G0/G1 cells represent apoptotic cells. Representative distributions of the cell cycle phases are depicted on day 0 of wild-type cells. (B) Cells were costained for propidium iodide to evaluate pre-G0/G1 apoptotic cells and for Ki-67 expression on nonapoptotic cells to evaluate proliferating cells. Cells were then analyzed by flow cytometry. Comparison of the mean fold increase in apoptosis versus mean fold decrease in proliferation of 4 independent experiments is shown. The values for apoptosis and proliferation were normalized to the value obtained for each day in control K562 cells, and assigned a value of 1.0 for proliferating cells and apoptotic cells. (C) Mobilized CD34+ cells from 2 donors for hematopoietic stem cell transplantation were purified, transduced, and induced to undergo erythroid differentiation as described in “Cell culture and erythroid differentiation.” After 5 days, cells were harvested and evaluated for apoptosis rates using annexin V staining. For each donor cell (N1 and N2), annexin V staining of cells transduced with scrambled RNA controls (SCR) or shRNA against SBDS (S-3) are shown. (D) Results of inhibition of SBDS mRNA in CD34+ cell samples are shown. The faster running SBDS band corresponds to a previously published shorter isoform. (Cell numbers did not allow corresponding testing of SBDS protein levels.)
To assess the effect of SBDS deficiency on proliferation during erythroid differentiation, cells were costained with propidium iodide and Ki-67.30 Viable (non–sub-G0/G1) cells were assessed for Ki-67 expression. Proliferation of SBDS-knockdown cells tended to be lower than that of the controls only in the cells with more severe SBDS-knockdown (K562/hSBDS-3; P = .05) compared with controls. Generally, the fold change in apoptosis was markedly higher than that in proliferation (Figure 3B). For example, on day 5 of differentiation, the increase in apoptosis in K562/shSBDS-3 cells was 3.6-fold compared with only a 1.29-fold decrease in proliferation. Because the percentage of cells at different stages of the cell cycle did not indicate accumulation at a specific phase (supplemental Figure 5), it is unlikely that SBDS-knockdown erythroid cells are arrested at a specific cell-cycle phase.
SBDS deficiency leads to increased levels of ROS during erythroid differentiation
We previously showed that SBDS-deficient HeLa and TF-1 cells have elevated ROS levels.16 It is unknown whether increased ROS levels have a detrimental role during differentiation of SBDS-deficient hematopoietic cells. Using 2′,7′-dichlorofluorescein-diacetate, which is oxidized to the fluorescent compound, dichlorofluorescein by intracellular H2O2, we found higher ROS levels in nondifferentiating SBDS-knockdown cells compared with controls. Importantly, the difference in ROS levels between SBDS-knockdown to control cells increased by 25% to 50% during differentiation (Figure 4A-C). In addition, we analyzed ROS levels in SBDS-knockdown CD34+ cells from 2 donors after 5 days in erythroid differentiation cultures. The SBDS-knockdown cells showed higher ROS levels than mock-transduced cells (Figure 4D-F).
Level of ROS. (A) For evaluation of intracellular ROS level, K562 cells (5 × 105) were washed and incubated for 30 minutes with 50M 2′,7′-dichlorofluorescein-diacetate (DCFH-DA). Cells and analyzed by flow cytometry. Duplicate cells were analyzed before inducing to undergo erythroid differentiation (a representative graph of 2 independent experiments). (B) Cells were analyzed 3 days after induction of erythroid differentiation (a representative graph of 2 independent experiments). (C) Calculation of the fold increase in the dichlorofluorescein fluorescence relative to the control wild-type cells. (D-E) Purified CD34+ from 2 donors were transduced with lentivectors containing either shSBDS-3 or shSCR control, sorted for expressing the YFP reporter gene, and induced erythroid differentiation. The cells were analyzed on day 5 for ROS levels by DCFH-DA. (F) Calculation of the fold increase in the dichlorofluorescein fluorescence relative to the scrambled RNA expressing cells.
Level of ROS. (A) For evaluation of intracellular ROS level, K562 cells (5 × 105) were washed and incubated for 30 minutes with 50M 2′,7′-dichlorofluorescein-diacetate (DCFH-DA). Cells and analyzed by flow cytometry. Duplicate cells were analyzed before inducing to undergo erythroid differentiation (a representative graph of 2 independent experiments). (B) Cells were analyzed 3 days after induction of erythroid differentiation (a representative graph of 2 independent experiments). (C) Calculation of the fold increase in the dichlorofluorescein fluorescence relative to the control wild-type cells. (D-E) Purified CD34+ from 2 donors were transduced with lentivectors containing either shSBDS-3 or shSCR control, sorted for expressing the YFP reporter gene, and induced erythroid differentiation. The cells were analyzed on day 5 for ROS levels by DCFH-DA. (F) Calculation of the fold increase in the dichlorofluorescein fluorescence relative to the scrambled RNA expressing cells.
Antioxidants improve the SBDS-deficient cell expansion capacity
To ask whether elevated ROS levels impair the expansion of SBDS-deficient cells, we performed rescue experiments with antioxidants. Addition of N-acetylcysteine to duplicate clonogenic assays of 3 SDS patients increased BFU-E colony numbers by 283%, 10%, and 40%, respectively (average 111%, P = .01). Addition of catalase to the 3 patients' cultures increased BFU-E colony numbers by 167%, 63%, and 27%, respectively (average 86%, P = .04; Figure 5A). The average increase in BFU-E colony production from CD34+ from 5 normal controls by N-acetylcysteine and catalase was 11% (P = .41) and 30% (P = .24), respectively. Addition of N-acetylcysteine and catalase to the SBDS-knockdown K562 cells, but not to controls, improved cell expansion (Figure 5B).
Rescue of SBDS-knockdown slow cell growth with antioxidants. (A) SDS patients and healthy control bone marrow CD34+ cells were plated in duplicates at a density of 1 × 103 cells/1 mL dish with serum-free containing methylcellulose and a cytokine cocktail (SCF, GM-CSF, IL-3, IL-6, G-CSF, and erythropoietin). Additional duplicate cultures were plated in the same conditions, but with N-acetylcysteine or catalase. BFU-E colonies containing 50 or more cells were scored after 14 days under an inverted microscope. (B) SBDS-knockdown and control K562 cells were induced to undergo erythroid differentiation by plating 2 × 104 cells/35-mm dishes with hemin. Replicate cultures were either not treated or treated with N-acetylcysteine 500μM or treated with catalase 500 U/mL and assessed daily for cell numbers by trypan blue exclusion. The cell growth of the SBDS-knockdown K562 cells after 5 days was significantly improved with N-acetylcysteine (P < .05) and catalase (P < .05) treatment.
Rescue of SBDS-knockdown slow cell growth with antioxidants. (A) SDS patients and healthy control bone marrow CD34+ cells were plated in duplicates at a density of 1 × 103 cells/1 mL dish with serum-free containing methylcellulose and a cytokine cocktail (SCF, GM-CSF, IL-3, IL-6, G-CSF, and erythropoietin). Additional duplicate cultures were plated in the same conditions, but with N-acetylcysteine or catalase. BFU-E colonies containing 50 or more cells were scored after 14 days under an inverted microscope. (B) SBDS-knockdown and control K562 cells were induced to undergo erythroid differentiation by plating 2 × 104 cells/35-mm dishes with hemin. Replicate cultures were either not treated or treated with N-acetylcysteine 500μM or treated with catalase 500 U/mL and assessed daily for cell numbers by trypan blue exclusion. The cell growth of the SBDS-knockdown K562 cells after 5 days was significantly improved with N-acetylcysteine (P < .05) and catalase (P < .05) treatment.
Ribosome biogenesis is more severely affected in differentiating erythroid cells
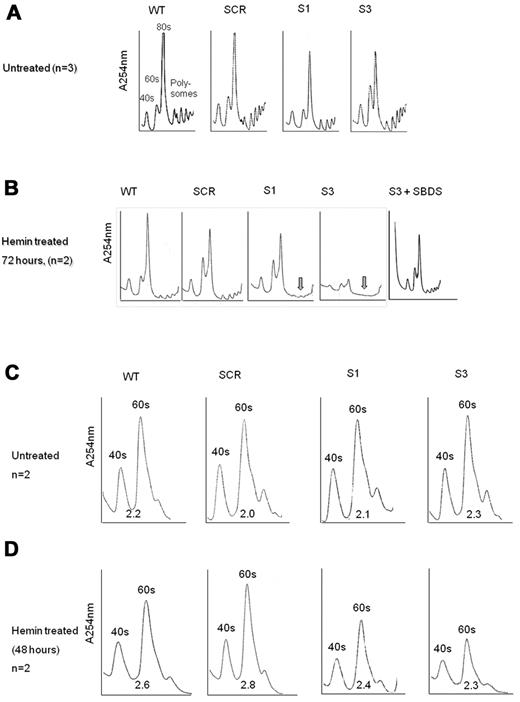
The apoptosis and proliferation data in Figure 3 support the notion that loss of SBDS during erythroid differentiation affects erythrocyte expansion, mainly by enhancing cell death. It is unknown, however, what events trigger apoptosis during differentiation of SBDS-deficient cells. Because SBDS plays a role in ribosome biogenesis18,31 and expanding cells require substantial amounts of energy, we hypothesized that defects in ribosome biogenesis would be exacerbated during erythroid cell development. We found that nondifferentiating SBDS-knockdown cells displayed slightly reduced amounts of 80S subunits compared with controls (Figure 6A). By contrast, during erythroid differentiation, SBDS-knockdown cells showed markedly impaired ribosome profiles with substantially reduced polysomes (Figure 6B). K562/shSBDS-3 cells, which express ∼ 5% of the normal SBDS protein concentration, also showed global reduction in all ribosome subunits, including 80S, 40S, and 60S subunits. Importantly, reintroduction of SBDS into SBDS-knockdown cells normalized the ribosome profile (Figure 6B), indicating that these alterations in ribosome profile are specifically the result of inhibition of SBDS. It is noteworthy that, after removal of dead cells, SBDS-knockdown cells still manifested general reduction of 80S subunits, indicating that this defect was not the result of accumulation of dead cells (supplemental Figure 6).
Ribosome profiles during differentiation of SBDS-knockdown cells. (A) Undifferentiated control and SBDS-knockdown K562 cells were lyzed using TMK100 buffer with cycloheximide. Equal amounts of RNA were layered on 5% to 45% sucrose gradients. Absorbance of each fraction at A254 was done by the Brandel Density Gradient Fractionator. The 40S, 60S, 80S, and polysomes peaks are seen on the graphs. A representative diagram of 3 independent experiments is shown. (B) SBDS-knockdown and control K562 cells were induced to undergo erythroid differentiation with hemin. After 72 hours, cells were lyzed and analyzed for ribosome profile as described in panel A. A representative diagram of 2 independent experiments is shown. Arrows are pointed to the markedly reduced polysomes in SBDS-knockdown cells. (C) The ribosomal profiles of undifferentiated control and SBDS-knockdown K562 cells were analyzed by sucrose gradient density centrifugation under conditions that allowed dissociation of the 40S and 60S subunits. Cells were harvested using TMK100 lysis buffer with cycloheximide and EDTA. Equal amounts of RNA were layered on 5% to 35% sucrose gradients. Absorbance of each fraction at A254 was done by the Brandel Density Gradient Fractionator. The ratios between 60S and 40S subunits were calculated by measuring area under the curve, using Adobe Illustrator, and are depicted under the curves. A representative diagram of 2 independent experiments is shown. (D) Ribosome profiles of differentiated K562 cells after 48 hours of hemin induction under EDTA dissociating conditions as described in panel C. A representative diagram of 2 independent experiments is shown.
Ribosome profiles during differentiation of SBDS-knockdown cells. (A) Undifferentiated control and SBDS-knockdown K562 cells were lyzed using TMK100 buffer with cycloheximide. Equal amounts of RNA were layered on 5% to 45% sucrose gradients. Absorbance of each fraction at A254 was done by the Brandel Density Gradient Fractionator. The 40S, 60S, 80S, and polysomes peaks are seen on the graphs. A representative diagram of 3 independent experiments is shown. (B) SBDS-knockdown and control K562 cells were induced to undergo erythroid differentiation with hemin. After 72 hours, cells were lyzed and analyzed for ribosome profile as described in panel A. A representative diagram of 2 independent experiments is shown. Arrows are pointed to the markedly reduced polysomes in SBDS-knockdown cells. (C) The ribosomal profiles of undifferentiated control and SBDS-knockdown K562 cells were analyzed by sucrose gradient density centrifugation under conditions that allowed dissociation of the 40S and 60S subunits. Cells were harvested using TMK100 lysis buffer with cycloheximide and EDTA. Equal amounts of RNA were layered on 5% to 35% sucrose gradients. Absorbance of each fraction at A254 was done by the Brandel Density Gradient Fractionator. The ratios between 60S and 40S subunits were calculated by measuring area under the curve, using Adobe Illustrator, and are depicted under the curves. A representative diagram of 2 independent experiments is shown. (D) Ribosome profiles of differentiated K562 cells after 48 hours of hemin induction under EDTA dissociating conditions as described in panel C. A representative diagram of 2 independent experiments is shown.
Because previous studies provided conflicting data on the ratio of 60S to 40S ribosome subunits in SBDS-deficient cells, we assayed the ribosome profile in the presence of EDTA. Normal levels and ratio of 40S and 60S subunits were detected in nondifferentiating SBDS-knockdown cells (Figure 6C). On differentiation, we detected a general reduction in both 40S and 60S subunits; however, the 40S/60S ratio was not affected (Figure 6D).
The impaired global translation of SBDS-deficient K562 is heightened during erythroid differentiation
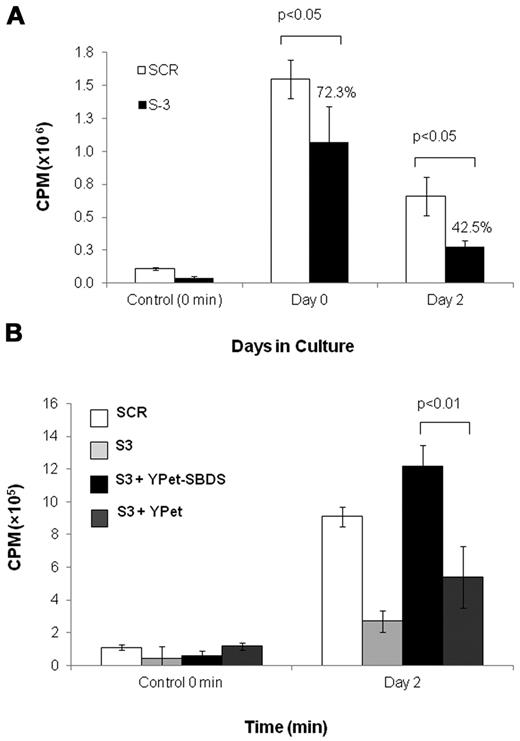
Next, we investigated whether the ribosome abnormalities found in SBDS-deficient cells resulted in reduced global translation. Global protein synthesis of nondifferentiating K562/shSBDS-3 cells was significantly reduced to 72% of K562/SCR cells (Figure 7A). After 48 hours of erythroid differentiation, the reduction of protein synthesis became more prominent as translation in K562/shSBDS-3 cells was reduced to 43% of K562/SCR cells (Figure 7A). Reintroduction of SBDS restored global translation during erythroid differentiation (Figure 7B). These results show that SBDS is necessary for normal protein translation, particularly during erythroid differentiation.
Global translation in SBDS-deficient cells. (A) Global translation in K562/shSBDS-3 and K562/SCR was studied by measuring incorporation of 35S-methionine/cysteine into equal amounts of newly synthesized trichloroacetic acid-precipitable peptides. Data are the mean ± SE of 3 experiments. Statistically significant results using t test: *P < .05. The ratios of the knockdown cells to control are presented in percentages. (B) Global translation in stable SBDS-knockdown cells after reintroduction of SBDS. Data are the mean ± SE of 3 experiments. *P < .05, statistically significant results using 1-way ANOVA and Tukey test for multiple means against both S-3 + YPET and S-3 cells.
Global translation in SBDS-deficient cells. (A) Global translation in K562/shSBDS-3 and K562/SCR was studied by measuring incorporation of 35S-methionine/cysteine into equal amounts of newly synthesized trichloroacetic acid-precipitable peptides. Data are the mean ± SE of 3 experiments. Statistically significant results using t test: *P < .05. The ratios of the knockdown cells to control are presented in percentages. (B) Global translation in stable SBDS-knockdown cells after reintroduction of SBDS. Data are the mean ± SE of 3 experiments. *P < .05, statistically significant results using 1-way ANOVA and Tukey test for multiple means against both S-3 + YPET and S-3 cells.
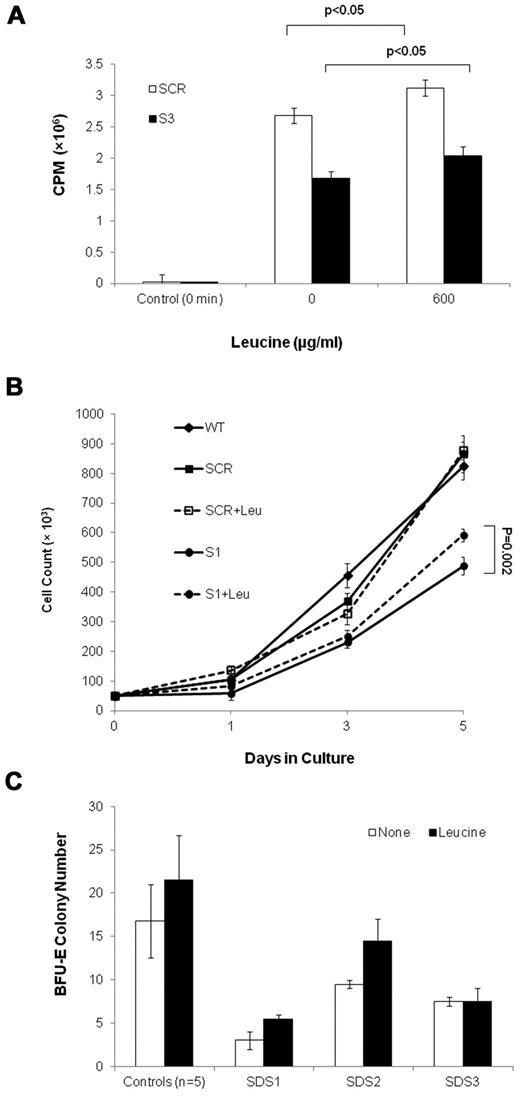
Leucine improves translation and cell expansion capacity of SBDS-deficient cells
To ask whether translation insufficiency has a detrimental effect on the expansion of SBDS-deficient cells, we performed rescue experiments in which a translation stimulator was added to marrow CD34+ cell cultures from 3 SDS patients. The addition of leucine, which activates the mTOR pathway and stimulates protein synthesis (Figure 8A),32-37 resulted in improved BFU-E colony growth in 2 of 3 patients, by 52% and 83%, respectively; whereas a third patient did not respond (Figure 8B). Addition of leucine to clonogenic assays of CD34+ cells from 5 normal controls resulted in an increase by an average of 28% (P = .25). In addition, leucine improved SBDS-deficient K562 cell expansion, which reaches a significant difference on day 5 after differentiation. Addition of leucine to K562/SCR (Figure 8C) and WT (data not shown) control cells did not increase cell numbers.
Leucine treatment of SBDS-deficient cells. (A) Translation in S-3 cells and SCR cells was studied by measuring incorporation of 35S-methionine/cysteine into equal amounts of newly synthesized trichloroacetic acid-precipitable peptides. Data are the mean ± SE of 3 experiments. *P < .05, statistically significant results (t test). (B) SBDS-knockdown and control K562 cells were induced to undergo erythroid differentiation by plating 2 × 104 cells/35-mm dishes with hemin. Replicate cultures were either treated or not treated with 600 μg/mL of leucine and assessed daily for cell numbers by trypan blue exclusion. The cell growth of the SBDS-knockdown K562 cells after 5 days was significantly improved with leucine treatment (P < .05). (C) SDS patients and healthy control bone marrow CD34+ cells were plated in duplicates at a density of 1 × 103 cells/1 mL dish with serum-free containing methylcellulose and a cytokine cocktail (SCF, GM-CSF, IL-3, IL-6, G-CSF, and erythropoietin). Additional duplicate cultures were plated in the same conditions, but with leucine. BFU-E colonies containing 50 cells or more were scored after 14 days under an inverted microscope.
Leucine treatment of SBDS-deficient cells. (A) Translation in S-3 cells and SCR cells was studied by measuring incorporation of 35S-methionine/cysteine into equal amounts of newly synthesized trichloroacetic acid-precipitable peptides. Data are the mean ± SE of 3 experiments. *P < .05, statistically significant results (t test). (B) SBDS-knockdown and control K562 cells were induced to undergo erythroid differentiation by plating 2 × 104 cells/35-mm dishes with hemin. Replicate cultures were either treated or not treated with 600 μg/mL of leucine and assessed daily for cell numbers by trypan blue exclusion. The cell growth of the SBDS-knockdown K562 cells after 5 days was significantly improved with leucine treatment (P < .05). (C) SDS patients and healthy control bone marrow CD34+ cells were plated in duplicates at a density of 1 × 103 cells/1 mL dish with serum-free containing methylcellulose and a cytokine cocktail (SCF, GM-CSF, IL-3, IL-6, G-CSF, and erythropoietin). Additional duplicate cultures were plated in the same conditions, but with leucine. BFU-E colonies containing 50 cells or more were scored after 14 days under an inverted microscope.
Genes that regulate ROS levels are differentially expressed in SDS marrow mononuclear cells
To determine whether the high ROS levels in SBDS-deficient erythroid cells are secondary to the defect in global translation, we treated SBDS-deficient K562 cells with leucine and evaluated ROS levels. Leucine did not reduce ROS levels as determined by the DCFH-DA assay at both 6 hours and 24 hours after administration on day 0, day 3, or day 4 of differentiation (data not shown). Thus, we analyzed the expression of genes involved in ROS regulation in SDS marrow mononuclear cells compared with controls. We found dysregulation of several genes, which may act together to increase ROS levels in SDS cells (supplemental Table 2). Nox4, which encodes the superoxide-generating enzyme NADPH-oxidase 4, was up-regulated in SDS cells. In addition, 3 genes that encode proteins that reduce ROS levels were down-regulated: EPX, CYGB, and FoxM1. EPX encodes eosinophil peroxidase, which catalyzes the conversion of H2O2 and bromide to hypobromite and water.38 CYGB encodes cytoglobin, which scavenges various ROS.39 FoxM1 encodes a transcription factor that up-regulates the expression of ROS scavenger genes.40
The p53 family members were not up-regulated in SBDS-deficient cells
Because p53 is activated in some disorders with defective ribosomes and translation,41-43 we asked whether the proapoptotic p53 family proteins were aberrantly expressed in SBDS-deficient cells. As previously reported,44 our K562 cells were deficient in p53 protein by Western blotting and its sequence showed biallelic nonsense mutations (data not shown). In addition, p63 protein was not detectable in K562 cells by Western blot analysis (data not shown). The proapoptotic p73 isoform TAp73 was expressed in K562 cells. TAp73 expression decreased during differentiation without significant difference between the SBDS knockdown and control cells (supplemental Figure 7A). We also analyzed the expression of p53, p63, and TAp73 in erythrocytes from BFU-E colony cells derived from CD34+ cells plated in the presence of erythropoietin and SCFs. After 14 days, only BFU-E colonies were observed in the clonogenic assays. The expression of p53 and TAp73 proteins in SDS patients' BFU-E colony cells was similar to that of the controls. p63 protein expression was slightly higher (by 15%-50%) in the SDS patients than the normal control cells (supplemental Figure 7B).
Discussion
Our findings suggest that SBDS is critical for normal erythropoiesis, and its deficiency impaired erythroid cell expansion. These data support that the anemia seen in most SDS patients is largely the result of primary bone marrow failure. In contrast to the impaired cell expansion of SBDS-deficient cells, erythroid differentiation program was maintained because hemoglobinization, TFR2 and GATA1 expression, and the proportion of normoblasts in patient marrows were not reduced. The mild elevation of pro-erythroblasts in patient bone marrows and the increase in hemoglobinization and GATA1 and TFR2 expression in K562 cells might reflect a positive feedback mechanism and stress erythropoiesis. Interestingly, previous studies using murine SBDS-deficient 32Dcl3 cells reported normal neutrophil differentiation.5 These data along with ours suggest that the hematopoietic defects in SDS are mainly the result of reduced expansion capacity of differentiating cells rather than a defect in the differentiation program itself. It is intriguing that a dramatic effect on global protein synthesis in SBDS-deficient cells had no negative effect on differentiation. This suggests that the balance between pro-differentiation and antidifferentiation factors allows differentiation in SBDS-deficient cells. ROS play a critical role in inducing exit of HSC/Ps from a quiescent state into differentiation.45 This raises the hypothesis that elevated ROS levels in SBDS-deficient cells play a role in this process.
For differentiation of K562 cells, we used hemin, which is known to increase hemoglobinization in normal erythroid precursors. Hemin was extensively used to study the erythroid differentiation program. It is a ferric chloride heme salt, which induces erythroid differentiation of HSC/Ps46 and erythroleukemia cell lines.47 The precise mechanism by which hemin induces differentiation is unclear, but it may involve modulating the activity of transcription factors, such as Oct-1 and GATA-1, or generating ROS, such as superoxide. When applying identical hemin concentration to the SBDS knockdown and control cells, we identified major differences between the cell types, suggesting a different erythrocyte response to differentiation inducers in the presence or absence of SBDS. Further, the aberrant expansion, survival, and oxidative stress of SBDS knockdown CD34+ HSC/Ps and the positive effect of antioxidants on SDS marrow cells suggest that the observed phenotype is not restricted to the usage of hemin and K562 cells.
We found that SBDS is up-regulated early during erythroid differentiation. SBDS expression in murine myeloid 32Dcl3 cells undergoing neutrophil differentiation also showed an early up-regulation. Interestingly, the expression pattern of the murine homologs of a few other inherited bone marrow failure genes, such as FANCC48 and RPS1949 followed a similar pattern. These data suggest a higher need for certain inherited bone marrow failure syndrome genes during early stages of hematopoietic cell development, when most of the expansion occurs. This is in contrast to the severe congenital neutropenia gene, ELA2, which is expressed mainly at the promyelocyte stage of neutrophils development.50
We showed that during erythroid differentiation SBDS deficiency leads to accelerated apoptosis. Because oxidative stress can activate apoptosis and because we have previously found high levels of ROS in SBDS-deficient cells, we asked whether high ROS levels impair the expansion of SBDS-deficient erythoid cells. The high ROS levels during differentiation and the ability of antioxidants to substantially improve cell expansion and colony formation of SBDS-deficient cells suggest that reduced expansion of differentiating erythroid cells is at least in part the result of high ROS levels. Furthermore, these ROS abnormalities may be the result of up-regulation of NOX4 and down-regulation of EPX, CYGB, and FOXM1 in these cells. Nox4 protein acts as an oxygen sensor and generates superoxide by transferring electrons from NADPH to molecular oxygen. Superoxide is converted to H2O2 by superoxide dismutase. Nox4 is expressed in various cells, including myeloid cells.51 In addition, 3 genes that are involved in ROS metabolism were down-regulated. EPX encodes eosinophil peroxidase, which metabolizes H2O2.38 It is expressed in various cells, including platelets.52 CYGB encodes cytoglobin, a ubiquitously expressed hexacoordinate hemoglobin that scavenges various ROS and provides a protective function during oxidative stress.39 FoxM1 encodes a transcription factor that up-regulates the expression of ROS scavenger genes, such as catalase.40 In future studies, the activity of these enzymes and factors in SBDS-deficient myeloid cells and the potential rescue by manipulating their levels have to be evaluated. It is interesting that apoptosis was accelerated in SBDS-knockdown K562 cells despite high GATA1 expression. Because apoptosis of SBDS-deficient marrow cells is mainly through the Fas pathway,10,11,13 it is possible that the positive effect of GATA1 on BCL-XL and cell survival was not apparent.
Loss of SBDS protein in differentiating erythroid cells resulted in a more severe reduction in all ribosome subunits, polysomes, and global translation than in nondifferentiating cells. SBDS deficiency has been suggested to interfere with TIF6 release from the 60S ribosomal subunit, thereby impairing its binding to 40S.18 Our results showing a reduced level of the mature ribosome subunits (80S) are consistent with these data. During differentiation, depletion of SBDS to very low levels by the shSBDS-3 cassette (∼ 5% of normal) resulted in reduced levels of 40S, 60S, 80S, and polysomes. This is consistent with the complex ribosome maturation defects observed in yeasts depleted of the SBDS homolog SDO1 by Moore et al.46 They showed a delay in 35S and 23S pre-rRNA processing, which suggests defects in cleavage of the 5′ end of 18S rRNA and in maturation of the 40S ribosome subunit. They also showed a delay in 27S rRNA processing, which suggests defects in cleavage between 5.8S and 25S and in maturation of the 60S ribosome subunit. The defect in K562/shSBDS-1 was also enhanced during differentiation, but to lesser degree than S3, which is probably the result of a higher level of SBDS (∼ 10%). Having a more severe phenotype during erythrocyte maturation suggests that SBDS regulation of ribosome biogenesis is particularly critical during differentiation and might be related to the fact that the hematopoietic system is specifically targeted in SDS. Erythrocytes have a particularly high demand for rRNA and protein synthesis during early stages of erythropoiesis. Therefore, deficiency of SBDS might limit the ability of the cells to meet this demand, thereby affecting their survival and expansion capacity. Because ribosome profile remained abnormal after removing dead cells, it is unlikely that the reduced ribosome biogenesis and translation are secondary to accelerated apoptosis. However, we cannot exclude the possibility that aberrant growth signal leads to the reduced ribosome biogenesis and translation.
An important unanswered question is whether abnormalities in ribosome biogenesis in SDS are the primary cause of the cell expansion defect. The protein synthesis stimulator leucine, improved colony production from SDS marrow CD34+ cells in 2 of 3 patients. Leucine significantly improved the expansion capacity of SBDS-knockdown K562 cells, but not of the controls. It is noteworthy that leucine did not completely abrogate the growth defects of SDS CD34+ cells and SBDS-knockdown K562 cells. Thus, is it probable that translation insufficiency contributes to, but is not the sole cause of, the hematopoietic failure in SDS. Alternatively, it is possible that more potent translation stimulators are necessary as leucine did not completely correct that global translation defect in SBDS-deficient cells. Because leucine did not improve ROS levels, it is probable that the mechanism of increased ROS levels is independent of the defect in translation insufficiency. Our gene expression data showing dysregulation of enzymes involved in ROS regulation are in agreement with this hypothesis.
Our analysis of human myeloid cells showed that SBDS deficiency leads to a similar reduction of all ribosome subunits. These data are consistent with results from human SBDS-deficient fibroblasts, where no reduction in the relative ratio between the 60S and 40S subunits was shown.19 Together with the results of our previous study showing reduced expression of multiple ribosomal protein genes of both the small and large ribosome subunits in SDS marrow cells,17 we conclude that SBDS is important for the integrity of all ribosome subunits by maintaining normal levels of its protein and rRNA components.
p53 has been suggested to mediate apoptosis in disorders of ribosome biogenesis.41-43,53 The role of p53 in causing expansion defects of maturing hematopoietic cells in SDS is unknown. The findings that K562 cells do not express p53 and that the expression of the protein in SDS erythroid colony cells was not higher than the controls suggest that the accelerated apoptosis in SBDS-deficient cells is independent of p53. The expression patterns of p73 and p63 in K562 cells and in patient erythroid cells suggest that also these genes are not related to accelerated apoptosis in SDS. Because p73 and p63 exist in multiple isoforms, our results do not exclude the possibility of pathogenetic significance of their particular isoforms. To further provide evidence of a lack of a role of the p53 family members in the reduced SDS hematopoietic cell expansion, it would be useful to inhibit the genes before induction of differentiation. However, the limited numbers of SDS HSC/Ps12 and their substantial sensitivity to in vitro manipulation render these experiments difficult to perform.
This study suggests that SBDS is critical for normal erythropoiesis. Translation is decreased, apoptosis and ROS levels are increased, and cell expansion is decreased during erythroid differentiation. We also showed that antioxidants and stimulators of protein synthesis improve the expansion capability of SBDS-deficient cells, providing the rationale for devising future preclinical and clinical studies to test the safety and efficacy of these agents as novel therapeutic strategies in SDS.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The work was supported by the Canadian Institute for Health Research (grant HPA161281) and Shwachman-Diamond Syndrome Canada. S.S. is a recipient of an Ontario Graduate Scholarship Award and the Hospital for Sick Children Research Competition Award.
Authorship
Contribution: S.S. performed research, analyzed data, and wrote the paper; H.W., C.L.N., K.Z., and J.Y. performed research and analyzed data; C.S.T. and M.S.I. performed research, contributed to research design, and wrote the paper; and Y.D. conceived and designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yigal Dror, Program in Cell Biology, Research Institute, and Marrow Failure and Myelodysplasia Program, Division of Haematology/Oncology, Department of Paediatrics, Hospital for Sick Children and the University of Toronto, 555 University Avenue, Toronto, ON M5G 1X8, Canada; e-mail: yigal.dror@sickkids.ca.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal