Abstract
Structural chromosomal rearrangements of the Nucleoporin 98 gene (NUP98), primarily balanced translocations and inversions, are associated with a wide array of hematopoietic malignancies. NUP98 is known to be fused to at least 28 different partner genes in patients with hematopoietic malignancies, including acute myeloid leukemia, chronic myeloid leukemia in blast crisis, myelodysplastic syndrome, acute lymphoblastic leukemia, and bilineage/biphenotypic leukemia. NUP98 gene fusions typically encode a fusion protein that retains the amino terminus of NUP98; in this context, it is important to note that several recent studies have demonstrated that the amino-terminal portion of NUP98 exhibits transcription activation potential. Approximately half of the NUP98 fusion partners encode homeodomain proteins, and at least 5 NUP98 fusions involve known histone-modifying genes. Several of the NUP98 fusions, including NUP98-homeobox (HOX)A9, NUP98-HOXD13, and NUP98-JARID1A, have been used to generate animal models of both lymphoid and myeloid malignancy; these models typically up-regulate HOXA cluster genes, including HOXA5, HOXA7, HOXA9, and HOXA10. In addition, several of the NUP98 fusion proteins have been shown to inhibit differentiation of hematopoietic precursors and to increase self-renewal of hematopoietic stem or progenitor cells, providing a potential mechanism for malignant transformation.
Introduction
One of the oldest, and most useful, whole genome screens for genes involved in malignant transformation is a simple karyotype of the malignant cell.1 Analysis of recurrent, nonrandom chromosomal translocation breakpoints has identified numerous genes important for malignant transformation and provided critical insight into the biology, classification, and prognosis of hematopoietic malignancies.2 The study of these genes (such as BCR-ABL and BCL2) has led to vastly improved therapy3 and has opened an entire field of scientific inquiry.4
The Nucleoporin 98 gene (NUP98) was originally identified as a structural component of the nuclear pore complex (NPC),5 and was subsequently shown to be a fusion partner with homeobox (HOX)A9 in acute myeloid leukemia (AML) patients with a t(7;11)(p15;p15).6,7 Twenty-eight distinct NUP98 gene fusions have been identified, caused primarily by balanced translocations and inversions, in the malignant cells of patients with a wide array of distinct hematopoietic malignancies, including AML, chronic myeloid leukemia in blast crisis (CML-bc), myelodysplastic syndrome (MDS), acute lymphoblastic leukemia (ALL), and bilineage/biphenotypic leukemia.8 In this overview, we present a summary of the known roles of NUP98 in normal cell physiology, the association of NUP98 fusion proteins with hematopoietic malignancies, the incidence and prognostic importance of these fusions, and the mechanisms by which NUP98 fusion oncoproteins contribute to the process of malignant transformation.
Normal functions of NUP98
NUP98 is a component of the NPC
NUP98 is an ∼ 90-kDa protein component of the NPC, a large multiprotein structure embedded in and traversing the nuclear membrane, and consists of ∼ 30 different proteins, many of which are present in multiple copies. NUP98 has been found on both the nucleoplasmic and cytoplasmic domains of the NPC.9-11 The NPC provides a bidirectional route of transport between the nucleus and the cytoplasm, allowing small ions and polypeptides to pass through by diffusion and larger macromolecules (mRNA and proteins > 40 kDa) by active transport mediated via carrier proteins and transport factors collectively called karyopherins (eg, importins, exportins, and transportin).
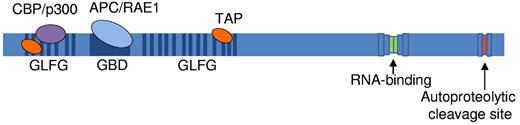
The NUP98 gene encodes 2 alternatively spliced mRNA variants: NUP98 and NUP98-NUP96. NUP98 and NUP98-NUP96 precursor polypeptides are cleaved into 90- (N-terminal) and 8- (C-terminal)-kDa peptides and 90- and 96-kDa peptides, respectively.5 The mature cleaved peptides are generated by NUP98 autoproteolytic cleavage12 that is necessary for correct targeting of NUP98 to the NPC5,9,13 (Figure 1).
Schematic representation of the NUP98 protein. Blue lines indicate GLFG repeats, the dark blue box indicates the GLEBS binding domain (GBD), and green and red boxes indicate the nucleoporin RNA-binding site and autoproteolytic cleavage site, respectively. Known NUP98 interacting factors are indicated; transcriptional coactivator CBP/p300 (purple) and the TAP transport cofactor (orange) bind the GLFG repeats. The APC/RAE1 complex (light blue) binds the GBD.
Schematic representation of the NUP98 protein. Blue lines indicate GLFG repeats, the dark blue box indicates the GLEBS binding domain (GBD), and green and red boxes indicate the nucleoporin RNA-binding site and autoproteolytic cleavage site, respectively. Known NUP98 interacting factors are indicated; transcriptional coactivator CBP/p300 (purple) and the TAP transport cofactor (orange) bind the GLFG repeats. The APC/RAE1 complex (light blue) binds the GBD.
Approximately a third of all nucleoporin proteins contain repeats of Phe-X-Phe-Gly amino acid residues, or Gly-Leu-Phe-Gly (GLFG) residues, collectively called FG repeats. However, NUP98 is distinct from other FG nucleoporins in that it contains multiple nontandem GLFG repeats.11 The GLFG repeats are thought to function as docking sites for karyopherins during trafficking of molecules through the NPC,11,14 and they have been shown to bind nuclear exportin 1 protein, XPO1 (the human homolog of yeast Crm1)15,16 and the mRNA export factor TAP.17
The nontandem FG repeats of NUP98 are intersected by a coiled-coil domain, the Gle2-binding sequence (GLEBS) motif in the N-terminal portion of NUP98 (Figure 1). The GLEBS motif binds the RNA export factor RAE1 (Gle2),18 and together the RAE1–NUP98 complex is capable of binding single-stranded RNA,19 whereas the FG repeats simultaneously bind TAP.17 The C-terminal end of NUP98 contains an RNA-binding motif.20-23 As precursor RNA molecules are spliced into their mature forms, they are packaged into messenger ribonucleoprotein particles consisting of these transport and export proteins and delivered to the NPC.
NUP98 also is found in the nucleoplasm and is involved in gene transcription
NUP98 was initially believed to function solely as a NPC component and chaperone in the transport and export of messenger ribonucleoprotein particles to the NPC, through the nuclear envelope, and into the cytoplasm. However, it has recently become evident that NUP98 has a broader role in normal cell physiology. NUP98 can be found diffusely throughout the nucleus, largely excluding the nucleolus, in focal clusters called “GLFG bodies.”15,16 The remarkable mobility of NUP98 seems to be dependent on active transcription by RNA polymerase, although exactly how remains unclear.15,24 Nup98 seems to function as a transport cofactor for the nuclear export protein Xpo1 (Crm1), interacting with Xpo1 via its GLFG repeats within the GFLG bodies in a Nup98-RanBP3-Xpo1-RanGTP-nuclear export signal cargo protein complex.
The NUP98 GLFG repeats bind CREB-binding protein (CBP)/p300, and a NUP98-GAL4 fusion protein was shown to have transcription activation potential, leading to the speculation that the amino-terminal portion of NUP98 contained a “cryptic” transactivation domain. Direct evidence for the participation of Nup98 in active transcription has recently been demonstrated in fruit flies. Discrete pools of Nup98 protein in the nucleoplasm or the NPC were identified in Drosophila cells,24,25 and the nucleoplasmic Nup98 was found to interact with actively transcribed genes bearing “active” chromatin marks such as H3K4me3, whereas the NPC-associated Nup98 did not associate with these chromatin marks.25 In addition, Nup98 was colocalized with RNA polymerase (pol) II chromatin “puff” domains24,25 and decreased levels of Nup98 protein resulted in a decrease in RNA pol II binding and decreased puff formation.24 Notably, although Nup98 mobility was found to be dependent on RNA pol II activity (active transcription), Nup98 binding of the target sites was not; therefore, Nup98 binding seems to precede active transcription and may play a role in induction of certain genes.24 Inhibition of Nup98 expression resulted in decreased expression of Nup98 target genes; similarly, overexpression of Nup98 led to increased target gene expression.25 Nup98 was found to bind specific chromatin sites, and Nup98 protein levels had the most effect on genes involved in cell cycle regulation and differentiation.24,25 In this context, it is interesting to note that expression of many NUP98 fusion genes impairs differentiation of hematopoietic precursors (see “A role for NUP98 in cell cycle progression and mitotic spindle formation”).
A role for NUP98 in cell cycle progression and mitotic spindle formation
In a complex with the Rae1 protein, Nup98 seems to be involved in mitotic spindle regulation. Dual haploinsufficiency of Nup98 and Rae1 has been shown to result in premature separation of sister chromatids, leading to severe aneuploidy.26,27 A chromosomal translocation that generates a fusion protein also results in loss of one NUP98 allele and therefore haploinsufficiency of NUP98, but not RAE1. However, if the NUP98 fusion protein acted to “sequester” RAE1 via binding to the GLEBS domain of NUP98, RAE1 levels would be functionally decreased, and the fusion protein could, in theory, confer haploinsufficiency of both NUP98 and RAE1. Although speculative, it is possible that aneuploidy resulting from NUP98 haploinsufficiency may be a mechanism whereby NUP98 fusion genes could cause genome instability, leading to acquisition of cooperating mutations, progression of disease, and clonal evolution.
Chromosomal translocations involving the NUP98 gene occur in a wide range of hematopoietic malignancies
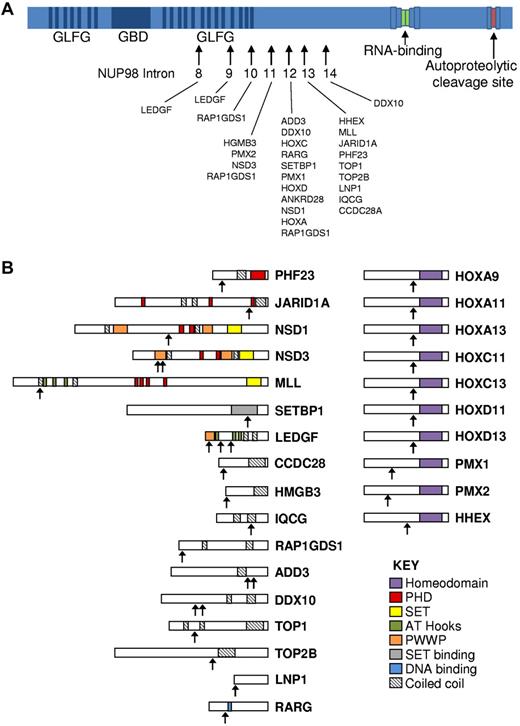
NUP98 was first linked to hematologic malignancies in 1996, with reports that the t(7;11)(p15;p15) translocation associated with AML generated a fusion gene that encoded the amino-terminal portion of NUP98 juxtaposed to the carboxyl-terminal portion of HOXA9.6,7 Subsequently, NUP98 was shown to fuse to numerous partner genes and is now known to produce abnormal fusion proteins with at least 28 different partner genes8 (Figure 2; Table 1).
NUP98 fusion proteins. (A) Schematic showing structure of the NUP98 protein and position of NUP98 fusion points in human leukemias. Arrows indicate fusion points. In all cases, the amino terminus of NUP98 is fused to the carboxyl terminus of the partner gene. (B) Schematic showing relevant domains of partner proteins and the position of the protein fusion. Domains are indicated in the key. Arrows indicate fusion point.
NUP98 fusion proteins. (A) Schematic showing structure of the NUP98 protein and position of NUP98 fusion points in human leukemias. Arrows indicate fusion points. In all cases, the amino terminus of NUP98 is fused to the carboxyl terminus of the partner gene. (B) Schematic showing relevant domains of partner proteins and the position of the protein fusion. Domains are indicated in the key. Arrows indicate fusion point.
NUP98 fusion partner genes
| Partner gene . | Chromosome . | Disease . | Relevant domain(s) . | Reference(s) . |
|---|---|---|---|---|
| HOXA9 | 7p15 | MDS, AML, CML, CMML | HD | 6,7 |
| HOXA11 | 7p15 | MDS, AML, CML, JMML | HD | 135 |
| HOXA13 | 7p15 | AML | HD | 136 |
| HOXC11 | 12q13 | AML | HD | 137 |
| HOXC13 | 12q13 | AML | HD | 138 |
| HOXD11 | 2q31 | AML | HD | 139 |
| HOXD13 | 2q31 | MDS, AML, CML | HD | 42 |
| PMX1 | 1q23 | AML, CML | HD | 140 |
| PMX2 | 9q34 | AML | HD | 141 |
| HHEX | 10q23 | AML | HD | 58 |
| PHF23 | 17p13 | AML | PHD, CC | 142 |
| JARID1A | 12p13 | AML | PHD, CC | 143 |
| NSD1 | 5q35 | MDS, AML, T-ALL | PHD, CC, SET | 144 |
| NSD3 | 8p11 | MDS, AML | PHD, CC, SET | 145 |
| MLL | 11q23 | AML | PHD, CC, SET | 121 |
| SETBP1 | 18q12 | T-ALL | CC | 35 |
| LEDGF | 9p22 | AML, CML | CC | 43 |
| CCDC28 | 6q24 | AML, T-ALL | CC | 146 |
| HMGB3 | Xq28 | AML | CC | 147 |
| IQCG | 3q29 | T-ALL | CC | 34 |
| RAP1GDS1 | 4q21 | AML, T-ALL | CC | 33 |
| ADD3 | 10q25 | AML, T-ALL | CC | 32 |
| DDX10 | 11q22 | MDS, AML, CML, CMML | CC | 148 |
| TOP1 | 20q11 | MDS, AML | CC | 149 |
| TOP2B | 3p24 | AML | CC | 150 |
| LNP1 | 3q12 | AML | CC | 151 |
| RARG | 12q13 | AML | CC | 36 |
| ANKRD28 | 3p25 | AML | Ankyrin | 152 |
| Partner gene . | Chromosome . | Disease . | Relevant domain(s) . | Reference(s) . |
|---|---|---|---|---|
| HOXA9 | 7p15 | MDS, AML, CML, CMML | HD | 6,7 |
| HOXA11 | 7p15 | MDS, AML, CML, JMML | HD | 135 |
| HOXA13 | 7p15 | AML | HD | 136 |
| HOXC11 | 12q13 | AML | HD | 137 |
| HOXC13 | 12q13 | AML | HD | 138 |
| HOXD11 | 2q31 | AML | HD | 139 |
| HOXD13 | 2q31 | MDS, AML, CML | HD | 42 |
| PMX1 | 1q23 | AML, CML | HD | 140 |
| PMX2 | 9q34 | AML | HD | 141 |
| HHEX | 10q23 | AML | HD | 58 |
| PHF23 | 17p13 | AML | PHD, CC | 142 |
| JARID1A | 12p13 | AML | PHD, CC | 143 |
| NSD1 | 5q35 | MDS, AML, T-ALL | PHD, CC, SET | 144 |
| NSD3 | 8p11 | MDS, AML | PHD, CC, SET | 145 |
| MLL | 11q23 | AML | PHD, CC, SET | 121 |
| SETBP1 | 18q12 | T-ALL | CC | 35 |
| LEDGF | 9p22 | AML, CML | CC | 43 |
| CCDC28 | 6q24 | AML, T-ALL | CC | 146 |
| HMGB3 | Xq28 | AML | CC | 147 |
| IQCG | 3q29 | T-ALL | CC | 34 |
| RAP1GDS1 | 4q21 | AML, T-ALL | CC | 33 |
| ADD3 | 10q25 | AML, T-ALL | CC | 32 |
| DDX10 | 11q22 | MDS, AML, CML, CMML | CC | 148 |
| TOP1 | 20q11 | MDS, AML | CC | 149 |
| TOP2B | 3p24 | AML | CC | 150 |
| LNP1 | 3q12 | AML | CC | 151 |
| RARG | 12q13 | AML | CC | 36 |
| ANKRD28 | 3p25 | AML | Ankyrin | 152 |
CMML indicates chronic myelomonocytic leukemia; JMML, juvenile myelomonocytic leukemia; and CC, coiled-coil.
It has been difficult to accurately estimate the incidence with which NUP98 fusions are associated with hematologic malignancy. One study of childhood AML patients from Austria identified one NUP98 rearrangement in 59 unselected cases,28 and a study designed to identify solely NUP98-HOXA9 fusions found 3 in 208 unselected cases of AML in an Asian population.29 The largest series identified 11 patients with NUP98-HOXA9 fusions among 493 consecutive AML cases in a Taiwanese population, for a frequency of 2.2%.30 There have been suggestions that NUP98-HOXA9 fusions are more common in Asian countries than Western countries, because Asian patients have been overrepresented in published series and case reports of NUP98-HOXA9 fusions.29,30 A study by the Groupe Francophone de Cytogenetique Hematologique demonstrated that 35% (23/66) of patients with hematopoietic malignancy and an 11p15 abnormality had a NUP98 translocation.31 Taken together, these data suggest that the frequency of NUP98 rearrangements in unselected patients with AML is 1% to 2% and that the frequency of NUP98 rearrangements in patients with an 11p15 abnormality is 35%.
NUP98 fusions are most common in myeloid malignancies, specifically AML, CML-bc, and MDS, but they have not been associated with myeloproliferative neoplasms (MPNs). A recent survey of 96 patients with NUP98 gene fusions indicated that patients with NUP98 fusions were relatively young (50% < 20 years) and that 25% of NUP98 fusions occurred in patients with therapy-related malignancy.31 Approximately 10% of patients with NUP98 fusions have T-lineage ALL (T-ALL); most commonly, these malignancies are associated with NUP98-RAP1GDS1 fusions. To date, no B-cell malignancies have been reported to bear a NUP98 fusion gene. Different partner genes are associated with different diseases, although such associations are rarely exclusive and are based on small numbers of patients. The NUP98-HOX gene fusions have not been observed in T-ALL patients and are limited to patients with MDS, AML, juvenile myelomonocytic leukemia, and chronic myelomonocytic leukemia. NUP98 fusions that have been associated with T-ALL include ADD332 , CCDC28A31 , RAP1GDS133 (each of which also have been associated with myeloid malignancies), IQCG,34 and SETBP1.35 Of note, the only reported case of a NUP98-RARG fusion was in a patient with promyelocytic leukemia,36 a leukemia that is almost always associated with a RARA fusion gene.37
Many NUP98 fusions are associated with overexpression of HOXA938,39 that in turn is associated with poor prognosis in AML.40,41 In addition, AML patients with a NUP98-HOXA9 fusion have worse overall survival and relapse-free survival.30 Therefore, although the patient numbers are small, NUP98 fusions seem to be associated with poor-prognosis AML.
NUP98 partner genes
All of the NUP98 gene fusions thus far identified encode a fusion protein that juxtaposes the amino-terminal portion of NUP98, containing the FG repeats, fused in frame to a partner gene, strongly suggesting that the oncogenic product is the 5′-NUP98-partner-3′ fusion. The reciprocal 5′-partner-NUP98-3′ fusion is often, but not invariably, expressed.42-44
NUP98 fusion partners can most simply be divided into 2 categories: homeodomain (HD) proteins and non-HD proteins. HD proteins can be further subdivided into clustered “class I” HD proteins (the HOX proteins) and “class II” nonclustered HD proteins. Ten HD proteins have been demonstrated to be involved in fusions with NUP98, including the most common and best studied of the fusion partners, HOXA9 (Figure 2; Table 1). The class II nonclustered HD genes involved in NUP98 fusions are HHEX, PMX1, and PMX2. In all instances, the C-terminal DNA-binding HD of the HD protein is retained in the fusion protein, and the transactivation domain is replaced by the GLFG repeats of NUP98.
Eighteen of the known NUP98 fusion partners do not encode HDs. Several of these genes lack a known DNA-binding domain, suggesting that they might have a mode of action distinct from that of the HD fusions. Although initially the non-HD partner genes were thought to have no common structural motifs, some patterns have emerged as more partner genes have been identified. The majority of these genes encode proteins that contain a coiled-coil domain that is thought to function in oligomerization of proteins (determinations made using the COILS 2.0 server45,46 ).
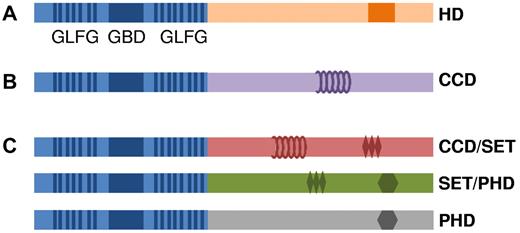
In addition to the coiled-coil domains, a recurring theme among some of the non-HD fusion partners is the presence of a histone “reading” or “writing” domain. In particular, 5 fusion partner genes encode plant HD (PHD) fingers, chromatin recognition domains that have been shown, in at least some cases, to be required for the leukemogenic potential of the fusion47 (Table 1). Three of these genes (NSD1, NSD3, and MLL) also encode supressor of variegation-enhancer of zeste-trithorax (SET) domains, which have a histone methyltransferase function. Indirectly, this theme is maintained for SETBP1 that interacts with a SET domain-containing protein. These proteins may comprise a third class of NUP98 fusion partner gene, with a specific mechanism of leukemogenesis (see “Proving oncogenicity: mouse models of NUP98 fusions”).
Proving oncogenicity: mouse models of NUP98 fusions
Mouse models of leukemia initiated by a wide range of leukemic fusion genes have been shown to recapitulate key features of the human malignancies in which they have been found, and they generally use 1 of 2 approaches. In the first, wild-type bone marrow cells transduced with a retrovirus encoding the leukemic fusion gene are transplanted into lethally irradiated recipients. An alternate approach is to generate genetically engineered (either transgenic or “knockin”) mice that express the fusion gene in the hematopoietic compartment. Both approaches have been used to study NUP98 fusion genes (Table 2).
Murine models of NUP98 fusions
| Fusion gene . | Phenotype . | Method . | Reference(s) . |
|---|---|---|---|
| NUP98-HOXA9 | MPN progressing to AML (mean latency, 230 d) | BMT | 48 |
| NUP98-HOXA9 | MPN progressing to AML (mean latency, 450 d) | Transgenic | 49 |
| NUP98-HOXA9 and Meis1 | MPN progressing to AML (mean latency, 142 d) | BMT | 48 |
| NUP98-HOXA9 and BCR-ABL | AML (latency, 21 d) | BMT | 123 |
| NUP98-HOXA9 and TEL-PDGFβR | AML (latency, 25 d) | BMT | 123 |
| NUP98-HOXA10 | MPN progressing to AML (mean latency, 223 d) | BMT | 50,51 |
| NUP98-HOXA10 and Meis1 | MPN progressing to AML (mean latency, 70 d) | BMT | 50,51 |
| NUP98-HOXD13 | MPN, not progressing to AML | BMT | 38 |
| NUP98-HOXD13 and Meis1 | MPN progressing to AML (median latency, 75 d) | BMT | 38 |
| NUP98-HOXD13 | MDS progressing to AML (median latency, 9 mo) | Transgenic | 54,55 |
| NUP98-PMX1 | MPN (mean latency, 246 d) | BMT | 57 |
| NUP98-PMX1 and Meis1 | MPN progressing to AML (median latency, 191 d) | BMT | 57 |
| NUP98-HHEX | AML (mean latency, 9 mo) | BMT | 58 |
| NUP98-TOP1 | AML (mean latency, 233 d) | BMT | 59 |
| NUP98-TOP1 and Meis1 | AML (mean latency, 242 d) | BMT | 59 |
| NUP98-NSD1 | AML (mean latency, 126 d) | BMT | 39 |
| NUP98-JARID1A | AML (mean latency, 69 d) | BMT | 47 |
| Fusion gene . | Phenotype . | Method . | Reference(s) . |
|---|---|---|---|
| NUP98-HOXA9 | MPN progressing to AML (mean latency, 230 d) | BMT | 48 |
| NUP98-HOXA9 | MPN progressing to AML (mean latency, 450 d) | Transgenic | 49 |
| NUP98-HOXA9 and Meis1 | MPN progressing to AML (mean latency, 142 d) | BMT | 48 |
| NUP98-HOXA9 and BCR-ABL | AML (latency, 21 d) | BMT | 123 |
| NUP98-HOXA9 and TEL-PDGFβR | AML (latency, 25 d) | BMT | 123 |
| NUP98-HOXA10 | MPN progressing to AML (mean latency, 223 d) | BMT | 50,51 |
| NUP98-HOXA10 and Meis1 | MPN progressing to AML (mean latency, 70 d) | BMT | 50,51 |
| NUP98-HOXD13 | MPN, not progressing to AML | BMT | 38 |
| NUP98-HOXD13 and Meis1 | MPN progressing to AML (median latency, 75 d) | BMT | 38 |
| NUP98-HOXD13 | MDS progressing to AML (median latency, 9 mo) | Transgenic | 54,55 |
| NUP98-PMX1 | MPN (mean latency, 246 d) | BMT | 57 |
| NUP98-PMX1 and Meis1 | MPN progressing to AML (median latency, 191 d) | BMT | 57 |
| NUP98-HHEX | AML (mean latency, 9 mo) | BMT | 58 |
| NUP98-TOP1 | AML (mean latency, 233 d) | BMT | 59 |
| NUP98-TOP1 and Meis1 | AML (mean latency, 242 d) | BMT | 59 |
| NUP98-NSD1 | AML (mean latency, 126 d) | BMT | 39 |
| NUP98-JARID1A | AML (mean latency, 69 d) | BMT | 47 |
BMT indicates retroviral bone marrow transduction.
NUP98-HOX fusions
Expression of the NUP98-HOXA9 fusion via bone marrow transduction and transplantation led to an MPN characterized by increased white blood cells, anemia, and decreased T- and B-cell progenitors.48 After a latency of at least 4 months, 11 of 14 mice progressed to AML. Secondary recipients demonstrated that the disease was transplantable, and coexpression of the Hox cofactor Meis1 reduced the latency to onset of AML. NUP98-HOXA9 transgenic mice showed comparable disease, with an MPN that transformed into AML in 22% of cases after a long latency of 15 months.49 Transduction of bone marrow cells with NUP98-HOXA10 produced a similar phenotype, with an MPN progressing to AML after a prolonged latency, that could be accelerated by cotransduction of Meis1.50,51 Surprisingly, coexpression of an Meis1 DNA-binding mutant also accelerated AML conversion, suggesting that the action of Meis1 may not be dependent on DNA binding.50,51
Expression of NUP98-HOXD13 by transduction of primary murine bone marrow cells also resulted in increased proliferation of immature myeloid cells in vitro.38 At 6 to 8 months after transplantation, several of the NUP98-HOXD13 recipients developed MPN, and several others developed severe anemia with T and B lymphopenia; coexpression of Meis1 led to rapid development of AML. Hoxa7 and Hoxa9 transcript levels were elevated in both the NUP98-HOXD13 and NUP98-HOXD13/Meis1 transplant recipients, although endogenous levels of Meis1 were not elevated.
Transgenic mice that expressed a NUP98-HOXD13 (NHD13) fusion under control of pan-hematopoietic Vav regulatory elements developed an MDS characterized by peripheral blood cytopenias, ineffective hematopoiesis, dysplasia, and apoptosis.52-54 Approximately 60% of the mice develop AML; the remaining mice died of severe pancytopenia or developed T-ALL or B-lineage ALL54,55 (S.M.G. and P.D.A., unpublished data, 2011). The lymphoid leukemias that developed in these mice demonstrated that expression of the NUP98-HOX fusions could be leukemic in lymphoid as well as myeloid cells. Bone marrow replating assays demonstrated increased repopulating abilities of the NHD13 marrow compared with wild-type marrow, similar to the findings with other NUP98-HOX fusions described under “NUP98-HOX fusions.” Lymphopoiesis in clinically healthy NHD13 mice was found to be partially blocked at the DN2-DN3 transition in thymocytes and at the pro-B to pre-B transition in B cells.56 Hoxa5, Hoxa7, Hoxa9, and Hoxa10 levels were markedly elevated in both the bone marrow and thymus; however, Meis1 mRNA levels were decreased compared with those found in wild-type bone marrow or thymus.53,56 Limiting dilution assays of NHD13 bone marrow identified the presence of a clonal transplantable MDS-initiating cell.53
Expression of a NUP98-PMX1 fusion resulted in MPN, with 50% of the mice dead by 300 days posttransplantation.57 NUP98-PMX1 enhanced myeloproliferation, impaired lymphopoiesis, and collaborated with Meis1 during AML transformation. These abilities were shown to be dependent on both the GLFG repeats of NUP98 and the DNA-binding activity of the PMX1 HD. Similar to the NUP98-HOX fusions, NUP98-PMX1 induced expression of the Hoxa cluster genes (Hoxa-5, -7, -9, and -10) that was dependent on the direct binding of the fusion protein to Hoxa regulatory regions.57
The NUP98-HHEX fusion also conferred aberrant self-renewal and impaired differentiation, properties that are dependent on the combined integrity of both the NUP98 FG repeats and the HD of HHEX.58 HHEX normally acts as a transcriptional repressor; however, its N-terminal repressive elements are replaced with the transactivating FG repeats of NUP98, transforming the HHEX repressive function into one of transcriptional activation. Bone marrow transplant of cells that expressed a NUP98-HHEX fusion resulted in AML that was associated with up-regulation of Hoxa5, Hoxa9, and Flt3.58
NUP98 non-HOX fusions
Expression of the NUP98-TOP1 fusion protein in a bone marrow transplant model led to MPN and AML.59 Similar to the NUP98-HOX fusions, both the NUP98 GLFG repeats and the TOP1 DNA-binding domain were essential for leukemogenesis; interestingly, leukemic transformation was independent of the topoisomerase activity of TOP1. In contrast to NUP98-HOX fusions, AML transformation was not accelerated by Meis1, suggesting the possibility that some aspect of transformation caused by the NUP98-TOP1 fusion is unique to this gene fusion.
Chromatin-modifying NUP98 fusions include NUP98-NSD1, NUP98-JARID1A, and NUP98-PHF23, and they may encode a distinct class of fusion oncoproteins. These fusions alter the conformation of chromatin by binding histone tails and “writing” or “reading” the active marks, thus modifying normal gene regulation. NUP98-NSD1–transduced bone marrow cells increased the expression of the Hoxa5, a7, a9, a10, and Meis1 and resulted in AML.39 NUP98-NSD1 was shown to bind and activate the promoters of Hoxa7 and Hoxa9 and was dependent on a PHD finger within the NSD1 portion of the fusion. Full transforming potential required the combination of the NUP98 GLFG repeats for recruitment of the CBP/p300 histone acetylation complex, the H3K36 methyltransferase activity of the NSD1 SET domain,60 and the binding of the Hoxa locus via the PHD domain. The NUP98-NSD1 fusion thus behaves as a potent transcriptional activator at the Hoxa locus by bringing together 2 epigenetic modifiers that prepare chromatin for active gene transcription, alongside a PHD domain that targets the Hoxa9 promoter.39 These findings support a model in which the NUP98-NSD1 fusion maintains a zone of active transcription by enforcing epigenetic modification at Hoxa gene regulatory elements while simultaneously preventing epigenetic silencing and repression by the polycomb repressor complex that normally silences the Hoxa locus during hematopoietic differentiation.
The epigenetic changes as just described also occur with NUP98-JARID1A and NUP98-PHF23 fusion proteins.47 Both fusions show transforming potential by enhancing progenitor cell self-renewal and arresting myeloblast differentiation. Mice transplanted with bone marrow cells transduced with the NUP98-JARID1A fusion developed AML, and transformation was shown to be dependent on the integrity of the PHD fingers that were determined to specifically bind di- or trimethylated H3K4 (H3K4me2/3). ChIP experiments showed binding of the NUP98-JARID1A fusion increased the expression of Hoxa5, a7, a9, and a10 genes. Mutation of conserved residues in the PHD fingers abrogated H3K4me3 binding, leading to failure of the fusion proteins to bind the Hoxa9 promoter, no change in Hoxa9 expression levels, and no leukemic transformation. Thus, the NUP98-JARID1A fusion represents a deregulated “reader” of histone marks and effectively blocks the epigenetic program required for normal cellular differentiation by maintaining expression of the Hoxa gene cluster.
Despite some degree of variation, potentially because of either the model system used or to the specific partner gene, several common themes can be observed from these in vivo models of NUP98 fusion genes. Anemia and aberrant differentiation of the myeloid lineage, with either increased or decreased circulating granuloctyes is present, and some of the models have reported impaired T- and B-cell differentiation. Marked overexpression of Hoxa cluster genes, most notably Hoxa5, a7, a9, and a10, is associated with the leukemic phenotype, and transformation to AML can be accelerated by coexpression of Meis1 in many cases. Both fusion partners possess protein domains essential for leukemogenesis, and collaborating mutations are likely to be required for evolution from a “preleukemic” MPN or MDS to AML.
Mechanism(s) of leukemic transformation initiated by NUP98 fusion genes
NUP98 fusion genes encode aberrant transcription factors
Several lines of evidence suggest that many of the NUP98 fusion proteins act as aberrant transcription factors. Although the wild-type NUP98 protein is primarily localized to the NPC, NUP98 fusion proteins are predominantly located in the nucleoplasm. The NUP98-HOXA9, NUP98-HOXD13, NUP98-TOP1, NUP98-PMX1, NUP98-NSD1, NUP98-IQCG, NUP98-PHF23, NUP98-JARID1A, NUP98-DDX10, and NUP98-HHEX fusion proteins have all been shown to be widely dispersed throughout the nucleus and in punctate foci called GLFG bodies in the nucleus, with the exception of the nucleoli.34,39,47,58,61-64 Although leukemogenicity has been shown to be dependent on the GLFG repeats that recruit the transcriptional coactivator complex CBP/p300,39,61,64,65 the interaction of NUP98 with RAE1 is not required for leukemic transformation.65
Furthermore, NUP98 fusion partner genes often contain domains known to be involved in gene transcription. Fusion partners seem to fall into 3 principal groups that either (1) directly bind DNA and activate gene transcription (HD transcription factors), (2) facilitate protein–protein interactions with transcriptional cofactors involved in active gene transcription (coiled-coil domains), or (3) modify chromatin to initiate gene transcription, maintain gene transcription, or both, gene transcription by “writing” or “reading” active chromatin marks (Figure 3). Coiled-coil domains have been shown to be necessary for the transforming abilities of the PML-RAR, AML1-ETO, and PAX-PML66,67 fusion proteins, and the proteins encoded by NUP98 partner genes DDX10, RAP1GDS1, LEDGF, TOP1, and NSD1 have been predicted or demonstrated45,68 to form coiled-coil motifs that can mediate protein–protein interactions.
Schematic representation of 3 archetypes of NUP98 fusions. (A) NUP98-HD fusion protein. (B) NUP98-coiled-coil motif fusion protein. (C) Three subgroups of NUP98 oncoproteins that potentially modify chromatin via coiled-coil domain (CCD), SET, or PHD finger motifs. The GBD is represented by a blue box, the GLFG repeats are shown as vertical blue lines, the HD domain is shown as an orange box, the CCD is shown as coils, the SET domain is shown as diamonds, and the PHD domain is shown as a hexagon.
Schematic representation of 3 archetypes of NUP98 fusions. (A) NUP98-HD fusion protein. (B) NUP98-coiled-coil motif fusion protein. (C) Three subgroups of NUP98 oncoproteins that potentially modify chromatin via coiled-coil domain (CCD), SET, or PHD finger motifs. The GBD is represented by a blue box, the GLFG repeats are shown as vertical blue lines, the HD domain is shown as an orange box, the CCD is shown as coils, the SET domain is shown as diamonds, and the PHD domain is shown as a hexagon.
Several NUP98 partner genes encode fusion proteins that contain PHD domains, SET domains, or both. As discussed already, the NUP98-NSD1 fusion protein has been shown to activate expression of Hoxa5, a7, a9, a10, and the Hox transcriptional cofactor Meis1.39 Chromatin modification was evident around the Hoxa7/a9 loci in cells that expressed a NUP98-NSD1 fusion compared with wild-type progenitor cells.39 In these cells, the Hoxa7/a9 locus was strongly H3 acetylated, displayed H3K36me3 marks, lacked H3K27me3 marks, and was bound by p300, all features that are consistent with active gene transcription.39 Three functionally distinct domains of the NUP98-NSD1 fusion protein were required for transcriptional activation of the Hoxa locus and immortalization of myeloid progenitor cells; the fifth PHD finger of NSD1, the SET domain of NSD1, and the NUP98 GLFG repeats. The NUP98-NSD1 fusion protein bound to chromatin 5′ of Hoxa7 and Hoxa9 via the PHD domain, recruited p300/CBP via the FG repeats, and methylated H3K36 via the SET domain, resulting in active gene transcription.39 Similar observations were made for the NUP98-JARID1A and NUP98-PHF23 fusions.47 In those studies, Hoxa5-a11 genes were overexpressed, and active H3K4me3 histone marks were localized to Hoxa5-a11, whereas flanking transcriptionally silent domains were marked by H3K27me and associated with the polycomb repressor complex components Suz12 and Ezh2.47 Deletion mutants of the PHD domains of JARID1A and PHF23 in the fusion proteins showed that the PHD fingers, which recognize H3K4me2/3, were essential for leukemic transformation. Binding of the PHD finger to H3K4me2/3 correlated with an open and active domain of gene transcription, suggesting that the fusion proteins prevented the spreading of H3K27me that is associated with silencing across the Hoxa locus during normal cell differentiation and maturation.
Given that some of the NUP98 fusions function as transcriptional regulators, several investigators have used gene expression arrays to identify genes and pathways regulated directly or indirectly by NUP98 fusion proteins. Analysis of NUP98-HOXA9 expression in human CD34+ cells using the Affymetrix U133+2.0 platform revealed up-regulation of interferon pathway genes (IFI44, IFI44L, IFIT1, MX1, and OAS1), HOX genes (HOXA3, HOXA5, and HOXA9), genes involved in myeloid differentiation (ELA2A and CTSG), and REN that encodes renin, a protease best known for its role in blood pressure regulation.69 A similar experiment using cord-blood CD34+ cells transduced with a NUP98-HOXA9 fusion and the Affymetrix U133A platform demonstrated up-regulation of HOX genes (HOXA5, HOXA6, HOXA7, HOXA9, and MEIS1), as well as RUNX1, CD44, PIM1, and HLF1.70 Expression of a NUP98-HOXA10 or NUP98-HOXD13 fusion in murine stem and progenitor cells led to overexpression of Crisp1, Nr4a1, Hlf1, Ahr, Pbx3, Hoxa5, and Hoxa7.71 Comparison of bone marrow from NUP98-HOXD13 transgenic mice to bone marrow from wild-type mice using a MEEBO/Stanford array demonstrated that 3 of the 10 most highly overexpressed genes were interferon-induced (Oas2, Ifit1, and Ifi44), and 4 encoded HD proteins (Hoxa7, Hoxa9, Hoxc6, and Pbx3; C.S. and P.D.A., unpublished data, 2007). However, in contrast to findings with other NUP98 fusion proteins, expression of the NUP98-HOXD13 fusion did not lead to up-regulation of Meis1 in that experiment. Expression of a NUP98-DDX10 fusion in human CD34+ cells demonstrated that the most up-regulated genes were HOXA cluster, MEIS1, and REN.63 Taken together, the data demonstrate that genes that encode HD proteins, particularly HOXA cluster genes, MEIS1 and PBX3, are typically up-regulated by expression of NUP98 fusion proteins, as are a group of interferon-inducible genes.
NUP98 fusion proteins deregulate HOXA cluster genes
The clustered HOX genes (class I HOX genes) are a family of genes that encode a conserved HD motif and function as DNA-binding transcription factors. The HOX genes were initially identified as “master regulators” of positional identity and body-pattern formation in Drosophila melanogaster (reviewed in McGinnis and Krumlauf72 ). Mammals have 39 HOX genes, organized in 4 discrete clusters designated A to D.73-75 A larger group of ∼160 less-conserved, nonclustered homeobox genes (class II HOX genes) are dispersed throughout the genome (reviewed in Argiropoulos and Humphries73 and Lawrence et al76 ).
Expression of Hoxa cluster genes is important for normal hematopoietic differentiation (reviewed in Argiropoulos and Humphries73 ). Although all 11 of the Hoxa cluster genes are expressed in murine HSCs, the most highly expressed and differentially regulated genes are Hoxa5, a7, a9, and a10.77 In general, these 4 Hoxa genes are most highly expressed in the hematopoietic stem and progenitor cells (HSPCs), and they seem to regulate HSPC self-renewal and repopulation of early myeloid and lymphoid progenitors.78-83 As progenitor cells proliferate and differentiate, thus losing their self-renewal properties, these 4 Hoxa cluster genes are down-regulated, as is the Hox cofactor Meis1.78,79,84
Approximately 50% of unselected human AML samples overexpress HOXA cluster genes, particularly HOXA7, A9, and A10, often in conjunction with the binding partner MEIS1.40,71,84-88 In addition, overexpression of HOXA9 has been linked to poor clinical outcome; indeed, a gene expression array study identified overexpression of HOXA9 as the single gene that most correlated with treatment failure in AML patients.41 Furthermore, several specific cytogenetic and molecular genetic AML subtypes overexpress HOXA cluster genes, including those with MLL-fusions,40,89,90 CALM-AF10 fusions,91 monosomy 7,92 and NPM1 mutants.93-95 Therefore, although NUP98 fusions are rare, they typically deregulate expression of HOXA cluster genes, particularly HOXA7, A9, and A10, whose overexpression is commonly associated with acute leukemia.
As we have described, expression of Meis1 was shown to enhance the leukemogenicity of NUP98 fusion proteins as well as Hoxa996 , and MEIS1 overexpression is often associated with overexpression of HOXA7 and HOXA9 in AML patients.71,86 MEIS1and the PBX1, PBX2, and PBX3 proteins are members of the “TALE” (3-amino acid loop extension) family of transcriptional cofactors that bind HOX proteins and serve to increase target specificity and stabilize DNA binding.97-101 Meis1 rather than the Pbx1 seems to be the more crucial cofactor for the transformation of myeloid bone marrow cells, because cotransduction of Meis1 and Hoxa9 resulted in AML in mice and cotransduction Pbx1 and Hoxa9 did not.102 A recent report indicates that levels of Meis1 may be the rate-limiting factor for leukemic transformation of MLL-fusion proteins, because higher levels of Meis1 correlated with reduced latency of disease.103 This also may be the case with NUP98-HOX fusions.
Of note, the microRNA (miR) miR-196b is located between HOXA9 and HOXA10 in both the mouse and human genomes. Small regulatory microRNAs play a major role in the regulation of gene expression (reviewed in Taft et al104 and Zhao et al105 ), and have now been profiled in leukemia samples to elucidate expression patterns that might signify specific genomic alterations, leukemia subtype classification, and treatment response.106-111 miR-196b is more highly expressed in progenitor than in differentiated hematopoietic cells, and it is thought to play a role in the regulation of normal hematopoiesis.112,113 Expression of miR-196b and HOXA9 and HOXA10 is highly correlated in leukemia samples,114 leading to the suggestion that miR-196b may be cotranscribed with the HOXA cluster.112,115 NUP98-HOXD13 and NUP98-PHF23 transgenic mice both overexpress miR-196b relative to wild-type bone marrow, and a transcript that contains both miR-196b and Hoxa9 sequences has been identified (S.M.G. and P.D.A., unpublished data, 2011), supporting the hypothesis that the Hoxa cluster is “open” to transcription in NUP98 fusion AMLs.
The mechanism(s) by which overexpression of HOXA cluster genes leads to leukemic transformation is unclear. However, given that expression of several HOXA cluster genes is down-regulated as stem cells mature, it is feasible that persistent expression of HOXA cluster genes enforces an immature stem or progenitor phenotype. HSC self-renewal is normally tightly controlled, and stem cells must balance self-renewal with generation of committed progenitors, so that sufficient numbers of committed progenitors are produced to sustain mature hematopoiesis. This balance is thought to be achieved through a combination of both asymmetric and symmetric HSC division, in which asymmetric division of HSCs gives rise to one committed daughter cell and one “self-renewal” cell. In contrast, symmetric division of HSCs gives rise to 2 self-renewing HSCs, leading to expansion of HSC number (reviewed in Morrison and Kimble116 ). Of note, although overexpression of BCR-ABL primarily increased the rate of hematopoietic precursor cell division, expression of NUP98-HOXA9 in murine cells led to increased symmetric division of hematopoietic precursors.117 In addition, enforced expression of a NUP98-HOXA9 fusion in human CD34+ cells led to expansion of HSCs, as measured by cobblestone formation and competitive repopulation assays.70 Taken together, these data indicate that expression of NUP98-HOXA9 can expand the pool of self-renewing, undifferentiated HSCs (Figure 4).
Simplified model of the relationship between Hoxa cluster gene expression and epigenetic modifications. (A) During normal hematopoiesis, Hoxa and miR-196b transcript levels are highest in HSPCs and decrease as cells differentiate to functional mature hematopoietic cells. In immature HSPCs, active histone marks such as H3K4me3 and the presence of trithorax group proteins correlate with high levels of Hoxa transcripts. These transcriptional enhancer marks decrease as cells mature and differentiate, and the Hoxa locus becomes progressively silenced by repressive H3K27me3 marks and polycomb repressor complex (PRC) group proteins (129,130-134; [130-134 are reviews]). (B) Mis-regulated Hoxa cluster gene expression associated with NUP98 fusion leukemogenesis, as a result of aberrant chromatin-modifying activities conferred by the NUP98 fusion.39,47 Mo indicates monocytes; Gran, granulocytes; Lym, lymphocytes; Ery, erythrocytes; Meg, megakaryocytes; and LICs, leukemia-initiating cells.
Simplified model of the relationship between Hoxa cluster gene expression and epigenetic modifications. (A) During normal hematopoiesis, Hoxa and miR-196b transcript levels are highest in HSPCs and decrease as cells differentiate to functional mature hematopoietic cells. In immature HSPCs, active histone marks such as H3K4me3 and the presence of trithorax group proteins correlate with high levels of Hoxa transcripts. These transcriptional enhancer marks decrease as cells mature and differentiate, and the Hoxa locus becomes progressively silenced by repressive H3K27me3 marks and polycomb repressor complex (PRC) group proteins (129,130-134; [130-134 are reviews]). (B) Mis-regulated Hoxa cluster gene expression associated with NUP98 fusion leukemogenesis, as a result of aberrant chromatin-modifying activities conferred by the NUP98 fusion.39,47 Mo indicates monocytes; Gran, granulocytes; Lym, lymphocytes; Ery, erythrocytes; Meg, megakaryocytes; and LICs, leukemia-initiating cells.
Events that collaborate with NUP98 fusion genes during leukemic transformation
It is generally accepted that mutations of a single gene are rarely sufficient to cause malignant transformation but that instead several collaborative mutations are required.118,119 A working framework for leukemic transformation has emerged in which one mutation from each of 2 distinct classes (“type I” and “type II”) is required.120 Type I mutations lead to increased proliferation, survival, or both, whereas type II mutations impair differentiation or enhance self-renewal of HSC/progenitor cells. The NUP98 fusions that have been studied seem to fall into the type II category, based on the ability of the NUP98-HOXD13 fusion54 to block 12-O-tetradecanoylphorbol-13-acetate–induced differentiation of K562 cells, and the impaired ability of HSCs and progenitor cells from NUP98-HOXD13 transgenic mice to differentiate in vitro.52 Recent work has indicated that this framework may be overly simplified, especially for the fusions such as NUP98-JARID1A, NUP98-PHF23, and NUP98-MLL that are known or suspected to function by modifying chromatin at numerous regions genome-wide,47,121 potentially affecting numerous genes with diverse functions.
The contention that some NUP98 fusions, such as the NUP98-HOX fusions, are type II mutations is supported by the identification of collaborating events in NUP98-HOX fusion leukemias, because most of the described collaborating events are type I proliferation and survival mutations. NUP98-HOXA9 fusions have been identified in patients with Ph+ CML-bc,122 and the collaborative nature of NUP98-HOXA9 and BCR-ABL fusions was confirmed in an experimental mouse model.123 Of 27 patients with NUP98 fusions (predominantly NUP98-HOX fusions), no mutations of MLL, NPM1, RUNX1, or CEBPA (all type II mutations) were identified; however, 25 type I events in total were found, including the FLT3-ITD mutation (12) and activating mutations in NRAS (5), KRAS (4), and KIT (4).30,124
A study of the complementary mutations that arise in leukemias of NUP98-HOXD13 transgenic mice supported the findings from human patients.125 In a screen of 32 leukemias, no type II mutations were identified (NpmI and Runx1 were screened), whereas almost one third of the mice had acquired a spontaneous type I mutation (4 Nras, 5 Kras, and 1 Cbl). None of the NUP98-HOXD13 mice with MDS that had not transformed to AML had evidence of a Nras or Kras mutation, suggesting that the Nras and Kras mutations are associated with progression of MDS to AML. Although Flt3 mutations did not occur spontaneously in that series, a recent study showed that the FLT3-ITD mutation markedly accelerates the onset of AML in NUP98-HOXD13 transgenic mice (S. Greenblatt and D. Small, personal e-mail communication, July 11, 2011), and another demonstrated that overexpression of the wild-type FLT3 accelerates the onset of AML in mice that express NUP98-HOXD13.126
Retroviral insertional mutagenesis has been used as a tool to identify collaborating mutations in several NUP98 fusion mouse models. One such study is a NUP98-HOXA9 model crossed onto a BXH2 background that identified cooperating events such as Meis1 and genes that encoded dynein motor complex proteins.49 A second mutagenesis screen performed on NUP98-HOXD13 mice identified Meis1 overexpression and Mn1 overexpression as important complementary events,127 and a third study identified inactivation of ICSBP as a complementary event for NUP98-TOP1 expression.128
Summary
The NUP98 protein has several distinct roles within the nucleus of the normal cell. Fusion of NUP98 to a large number of “partner genes” leads to the generation of leukemogenic NUP98 fusion proteins. NUP98 fusions are associated with a wide spectrum of hematopoietic malignancies, including MDS, AML, CML-bc, and pre-T lymphoblastic lymphoma. It is not yet clear whether this diversity reflects the NUP98-partner gene, the cell type that undergoes the initial NUP98 fusion event, or a combination of both. Complementary mutations frequently recognized in NUP98 fusion leukemias include genes that encode proteins involved in proliferation, resistance to apoptosis, or both, such as BCR-ABL, NRAS, KRAS, and FLT3. A common theme that emerges among many of the leukemias that express NUP98 fusion proteins is deregulation of the HOXA gene cluster, leading to impaired terminal differentiation of hematopoietic cells. The fact that deregulation of HOXA cluster gene expression is a frequent finding in leukemias initiated by NUP98 fusions make this cluster of genes an attractive target for novel therapeutic approaches.
Acknowledgments
The authors apologize to those whose work was not cited because of space limitations. They thank R. Keith Humphries, Guy Sauvegeau, Nicolas Pineault, Trang Hoang, Michael Heuser, David Curtis, Glenn Begley, Ilan Kirsch, Michael Kuehl, Paul Meltzer, Kevin Gardner, Kenneth Gross, Sheila Jani-Sait, Michael Thirman, Stephen Nimer, Kevin Gardner, J. P. Issa, Eley Estey, Guillermo Garcia-Manero, Donald Small, Sarah Greenblatt, Terry Fry, Stephen Jane, Alex Dobrovic, and past and present members of the Aplan laboratory for many fruitful discussions.
This research was supported by the Intramural Research Program, National Cancer Institute, National Institutes of Health; the Cancer Council of Victoria; and the National Health and Medical Research Council of Australia (grant 628367).
National Institutes of Health
Authorship
Contribution: S.M.G. and C.I.S. wrote the first draft of the review; and P.D.A. edited the final draft.
Conflict-of-interest disclosure: P.D.A. and C.I.S. receive royalties from the National Institutes of Health Technology Transfer office for the invention of NUP98-HOXD13 mice. The remaining author declares no competing financial interest.
Correspondence: Peter D. Aplan, Genetics Branch, Center for Cancer Research, National Cancer Institute, National Institutes of Health, 41 Center Dr, Bethesda, MD 20892; e-mail: aplanp@mail.nih.gov.

![Figure 4. Simplified model of the relationship between Hoxa cluster gene expression and epigenetic modifications. (A) During normal hematopoiesis, Hoxa and miR-196b transcript levels are highest in HSPCs and decrease as cells differentiate to functional mature hematopoietic cells. In immature HSPCs, active histone marks such as H3K4me3 and the presence of trithorax group proteins correlate with high levels of Hoxa transcripts. These transcriptional enhancer marks decrease as cells mature and differentiate, and the Hoxa locus becomes progressively silenced by repressive H3K27me3 marks and polycomb repressor complex (PRC) group proteins (129,130-134; [130-134 are reviews]). (B) Mis-regulated Hoxa cluster gene expression associated with NUP98 fusion leukemogenesis, as a result of aberrant chromatin-modifying activities conferred by the NUP98 fusion.39,47 Mo indicates monocytes; Gran, granulocytes; Lym, lymphocytes; Ery, erythrocytes; Meg, megakaryocytes; and LICs, leukemia-initiating cells.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/24/10.1182_blood-2011-07-328880/4/m_zh89991181740004.jpeg?Expires=1765001871&Signature=KYmkwgy3ex2QUI7zDSxKLZDpPt2RjAd6bjlNfsLaK5g-fTYtJ7D2rJ4OrNY46nLbJE7gscINgr0Vr7LBSZeXM3TP2eyRuA8VlsucRoXDpN6a61BIWWDus7uFFVcyp5Evpnix7xHGPC1xJi7fzji~GQkM7Klo-euNdeMfvjJMNPfoGEQkQwqLbwLhjCkpNskrPvSS53Gpg28xlLcdmO1C7jjUIMbD1KiOX5Ty3eT8ntzcUIutghK4AhVZKFyfT2Sipq5km344OJfwJusTPVJ7Nd1lo1YMAXwg5vYy9DROM8WvpWxjkG8i2zAgN3HaGps~eQL6Fk-c--0nJJ0KPtGvhQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)