Abstract
To further our understanding of the genetic basis of acute myelogenous leukemia (AML), we determined the coding exon sequences of ∼ 18 000 protein-encoding genes in 8 patients with secondary AML. Here we report the discovery of novel somatic mutations in the transcriptional corepressor gene BCORL1 that is located on the X-chromosome. Analysis of BCORL1 in an unselected cohort of 173 AML patients identified a total of 10 mutated cases (6%) with BCORL1 mutations, whereas analysis of 19 AML cell lines uncovered 4 (21%) BCORL1 mutated cell lines. The majority (87%) of the mutations in BCORL1 were predicted to inactivate the gene product as a result of nonsense mutations, splice site mutation, or out-of-frame insertions or deletions. These results indicate that BCORL1 by genetic criteria is a novel candidate tumor suppressor gene, joining the growing list of genes recurrently mutated in AML.
Introduction
Acute myeloid leukemia (AML) will occur in approximately 13 000 new cases per year in the United States and result in 9000 untimely deaths. Recurrent chromosomal alterations provided some of the first clues to the pathogenesis of AML and contributed to improved classification of its subtypes. The molecular characterization of these translocations, along with identification of genes harboring intragenic mutations, has further elucidated disease mechanisms and provided new diagnostic and therapeutic approaches.1,2 These genes include FLT3, NPM1, CEBPA, TP53, IDH1 and IDH2, TET2, WT1, RUNX1, ASXL1, EZH2, NF1, and DNMT3A.2-18 Historically, most mutated genes driving cancers have been thought to regulate specific, well-defined pathways, such as those associated with RAS/RAF, PTEN/PIK3CA, and APC/β-catenin.19 More recently, unbiased genome-wide studies have identified mutated genes that appear to deregulate global patterns of gene expression through effects on chromatin structure or DNA methylation (eg, IDH1/2, TET2, EZH2, and DNMT3A).20-24
To extend our knowledge of subtle genetic alterations involved in AML, we have studied 8 secondary AML cases by exomic sequencing and have extended findings into a well-characterized unselected cohort of 173 AML cases as well as 19 AML cell lines. Through these efforts, we have identified novel somatically acquired inactivating mutations in the transcriptional corepressor gene BCORL1 in AML.
Methods
Patients and samples
The AML patients studied in this report were consecutively enrolled between March 2005 and October 2010 at the University of Michigan Comprehensive Cancer Center. The study was approved by the University of Michigan Institutional Review Board (IRBMED #2004-1022), and written informed consent was obtained from all patients before enrollment in accordance with the Declaration of Helsinki. Clinical and other characteristics of the 8 secondary AML cases used in the discovery screen are detailed in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). For the prevalence screen, 165 additional unselected AML cases (exclusive of AML-FAB-M3) were analyzed; the clinical characteristics of this AML cohort are detailed in supplemental Table 2. (For all other methods, see supplemental Methods.)
Results
Patient characteristics
Characteristics of the 173 AML patients (all non-M3 French-American-British subtype) analyzed in this study are summarized in supplemental Tables 1 to 3. Of these patients, 58% had de novo AML, 33% had AML with myelodysplasia-related changes (World Health Organization 2008 MRC1-3), and 9% had treatment-related AML (the latter category does not include patients that received MDS-directed therapies only). Eighty percent of patients had not been treated for AML at trial enrollment, whereas 20% had been treated for AML and relapsed at the time their samples were obtained.
Discovery screen
We determined the sequences of approximately 18 000 protein-encoding genes in 8 patients with secondary AML (supplemental Table 1). Massively parallel sequencing of captured tumor and matched normal DNA resulted in an average depth of coverage of each base in the targeted regions of 92-fold; 94.1% of the bases were represented by at least 10 reads (supplemental Table 4).
Using stringent criteria, we found 5 genes to be somatically mutated in 2 tumors in the discovery set, including BCORL1, NRAS, IDH1, DNMT3A, and RUNX1. We also identified one IDH2 mutation and one TP53 mutation in 1 of 8 discovery tumors.
Prevalence screen
Based on the results of the discovery screen, we initially evaluated the BCORL1, NRAS, IDH1, IDH2, DNMT3A, RUNX1, TP53, FLT3, and NPM1 genes for mutations in an additional 87 AML cases that were intentionally enriched for secondary AML (AML with myelodysplasia-related changes; supplemental Table 5). In each case, we amplified each exon of each gene from purified blast cell DNA and used conventional Sanger sequencing to search for mutations. Any mutations identified in the tumors were tested to determine whether they were somatic by similar sequencing of paired normal DNA from the same patients. Through these efforts, we identified 6 additional cases of AML with BCORL1 mutations. Patient AML 35 was preceded by CMML1. We tested 2 consecutively procured bone marrow samples from patient 35 in the CMML1 phase of the illness and detected the BCORL1 mutation at both times.
In addition to BCORL1, several of the other tested genes had mutations in these additional 87 cases (supplemental Tables 6 and 7). Whereas mutations in FLT3 and NPM1 were more prevalent in primary compared with secondary AML, mutations in BCORL1, DNMT3A, RUNX1, IDH1, and IDH2 were equally distributed. As expected, TP53 mutations were more common in secondary and t-AML than in de novo AML.
Next, we evaluated the BCORL1 gene in an additional 78 AML cases, thus complementing the analysis for this gene for all AML in our consecutively enrolled AML cohort (N = 173). Two additional BCORL1 mutated cases were identified, bringing the total to 10 (of 173 cases analyzed) and providing a BCORL1 mutation prevalence estimate of 5.8%. Characteristics of the 10 AML cases with BCORL1 mutations are summarized in Table 1. None of the BCORL1 mutated cases was TP53, CEBPA, or NPM1 mutated.
Characteristics of AML cases with BCORL1 mutations
| Sample . | Age, y . | Sex . | Previous treatment for AML . | FAB subclass . | WHO type . | Prior MDS or MDS/MPN . | Therapy-related . | SWOG risk group . | Cytogenetic risk . | BCORL1 mutations . | KRAS mutations . | NRAS mutations . | CEBPA mutations . | IDH1 mutations . | IDH2 mutations . | DNMT3A mutations . | TP53 mutations . | RUNX1 mutations . | FLT3 mutations . | NPM1 mutations . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AML14 | 70 | Female | No | M1 | AML-NOS | No | No | Intermediate | 47,XX,+8[19] | p.S1158A | WT | WT | WT | WT | p.R140Q | WT | WT | Frameshift | p.D835Y | WT |
| AML151 | 51 | Male | No | M4 | AML-MRC-1,2 | RAEB-1 | No | Unfavorable | 45,XY,-7,i(21)(q10)[12]/46,XY, +i(21)(q10)[6] | p.P810T | WT | p.Q61K | WT | WT | WT | WT | WT | p.W106L | WT | WT |
| AML33 | 68 | Male | No | M6 | AML-MRC-1 | RAEB-2 | No | Intermediate | 46,XY,del(3)(q13q27)[16]/46,XY[4] | Frameshift | WT | WT | WT | WT | WT | p.V716I | WT | WT | WT | WT |
| AML35 | 67 | Male | No | M2 | AML-MRC-1 | CMML-1 | No | Intermediate | 47,XY,+14[16]/46,XY[4] | Frameshift | WT | WT | WT | WT | WT | p.Y448X | WT | WT | WT | WT |
| AML45 | 63 | Female | No | M4 | AML-TR | No | Yes | Intermediate | 46,XY[20] | Frameshift | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT |
| AML216 | 50 | Female | No | M5 | AML-MRC-1 | RCMD | No | Intermediate | 47,XX,+8[5]/46,XX[24] | Frameshift | WT | WT | WT | p.R132C | WT | p.S714F, p.G685R | WT | WT | WT | WT |
| AML257 | 63 | Female | No | M4 | AML-NOS | No | No | Intermediate | 46,XX[20] | Frameshift | WT | WT | WT | WT | WT | WT | WT | Frameshift; p.F131V | ITD | WT |
| AML57 | 69 | Male | No | M4eo | AML-RGA (16p13q22) | No | No | Favorable | 46,XY,inv(16)(p13q22)[14] | p.Q538X | p.G13GD | WT | WT | WT | WT | WT | WT | WT | WT | WT |
| AML87 | 55 | Female | No | M0 | AML-NOS | No | No | Unfavorable | 46,XX,t(3;9;22)(q21;q34;q11.2) [15]/46,XX[5] | p.E1112X | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT |
| AML182 | 59 | Male | No | M0 | AML-NOS | No | No | Intermediate | 48,XY,+21,+21[15] | p.R784X | WT | WT | WT | p.R132G | WT | p.R693H | WT | Frameshift | ITD | WT |
| Sample . | Age, y . | Sex . | Previous treatment for AML . | FAB subclass . | WHO type . | Prior MDS or MDS/MPN . | Therapy-related . | SWOG risk group . | Cytogenetic risk . | BCORL1 mutations . | KRAS mutations . | NRAS mutations . | CEBPA mutations . | IDH1 mutations . | IDH2 mutations . | DNMT3A mutations . | TP53 mutations . | RUNX1 mutations . | FLT3 mutations . | NPM1 mutations . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AML14 | 70 | Female | No | M1 | AML-NOS | No | No | Intermediate | 47,XX,+8[19] | p.S1158A | WT | WT | WT | WT | p.R140Q | WT | WT | Frameshift | p.D835Y | WT |
| AML151 | 51 | Male | No | M4 | AML-MRC-1,2 | RAEB-1 | No | Unfavorable | 45,XY,-7,i(21)(q10)[12]/46,XY, +i(21)(q10)[6] | p.P810T | WT | p.Q61K | WT | WT | WT | WT | WT | p.W106L | WT | WT |
| AML33 | 68 | Male | No | M6 | AML-MRC-1 | RAEB-2 | No | Intermediate | 46,XY,del(3)(q13q27)[16]/46,XY[4] | Frameshift | WT | WT | WT | WT | WT | p.V716I | WT | WT | WT | WT |
| AML35 | 67 | Male | No | M2 | AML-MRC-1 | CMML-1 | No | Intermediate | 47,XY,+14[16]/46,XY[4] | Frameshift | WT | WT | WT | WT | WT | p.Y448X | WT | WT | WT | WT |
| AML45 | 63 | Female | No | M4 | AML-TR | No | Yes | Intermediate | 46,XY[20] | Frameshift | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT |
| AML216 | 50 | Female | No | M5 | AML-MRC-1 | RCMD | No | Intermediate | 47,XX,+8[5]/46,XX[24] | Frameshift | WT | WT | WT | p.R132C | WT | p.S714F, p.G685R | WT | WT | WT | WT |
| AML257 | 63 | Female | No | M4 | AML-NOS | No | No | Intermediate | 46,XX[20] | Frameshift | WT | WT | WT | WT | WT | WT | WT | Frameshift; p.F131V | ITD | WT |
| AML57 | 69 | Male | No | M4eo | AML-RGA (16p13q22) | No | No | Favorable | 46,XY,inv(16)(p13q22)[14] | p.Q538X | p.G13GD | WT | WT | WT | WT | WT | WT | WT | WT | WT |
| AML87 | 55 | Female | No | M0 | AML-NOS | No | No | Unfavorable | 46,XX,t(3;9;22)(q21;q34;q11.2) [15]/46,XX[5] | p.E1112X | WT | WT | WT | WT | WT | WT | WT | WT | WT | WT |
| AML182 | 59 | Male | No | M0 | AML-NOS | No | No | Intermediate | 48,XY,+21,+21[15] | p.R784X | WT | WT | WT | p.R132G | WT | p.R693H | WT | Frameshift | ITD | WT |
FAB indicates French-American-British; WHO, World Health Organization; MDS, myelodysplastic syndrome; MPN, myeloproliferative neoplasm; SWOG, Southwest Oncology Group; NOS, not otherwise specified; MRC, Medical Research Council; RAEB, refractory anemia with excess blasts; CMML, chronic myelomonocytic leukemia; RCMD, refractory cytopenia with multilineage dysplasia; and WT, wild-type.
The coding exons of the BCORL1 gene were also sequenced in 24 cell lines, of which 19 were AML-derived. In total, 5 BCORL1 sequence variants were identified, including 3 nonsense, 1 frameshift, and 1 splice junction mutation (located in the conserved nucleotide immediately preceding exon 10). Data are summarized in supplemental Table 8.
BCORL1 is a transcriptional corepressor encoding a protein that is found tethered to promoter regions by DNA-binding proteins. BCORL1 contains a putative bipartite nuclear localization signal, tandem ankyrin repeats, and a classic C-terminal binding protein (CtBP)–binding motif, PXDLS. Functional studies25 have shown that BCORL1 can interact with class II histone deacetylases, interact with the CtBP corepressor through the CtBP binding motif-PXDLS and affects the repression of E-cadherin.25
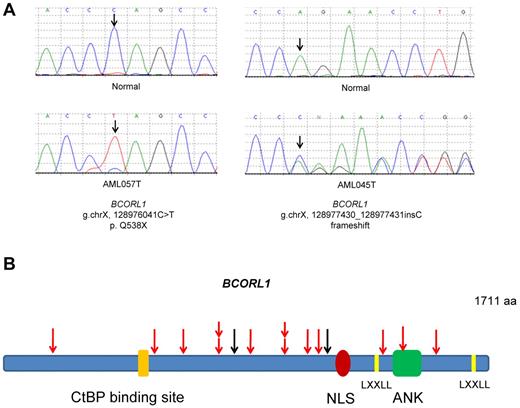
The BCORL1 gene is located on the X-chromosome at cytoband Xq25-q26.1 at approximately physical position 129.14 to 129.19 Mb. Eight of 10 (80%) BCORL1 mutations found in primary cells and all BCORL1 mutations in cell lines truncated the encoded protein as a result of nonsense mutations, splice site mutation, or out-of-frame insertions or deletions. All the truncating mutations in BCORL1 were predicted to result in severely shortened polypeptides lacking the LXXLL nuclear receptor recruitment motif and the C-terminus (Figure 1). These data not only provide strong evidence that BCORL1 mutations are important to the pathogenesis of AML but demonstrate that BCORL1 is, on the basis of genetic criteria, a tumor suppressor gene that is inactivated by mutations.
Schematic representations of the location of somatic mutations identified in the BCORL1 gene. (A) Examples of sequence chromatograms showing inactivating somatic mutations of BCORL1 in blast cells (bottom panels) but not in their matched normal buccal mucosa (top panels). (B) Schematic of somatic mutations identified in the BCORL1 gene in primary AML cells and AML cell lines. Red arrows indicate truncating mutations; and black arrows, missense mutations. NLS indicates nuclear localization signal; LXXLL, Leu-Xaa-Xaa-Leu-Leu motif; ANK, tandem ankyrin repeats; aa, amino acids; and CtBP, C-terminal binding protein.
Schematic representations of the location of somatic mutations identified in the BCORL1 gene. (A) Examples of sequence chromatograms showing inactivating somatic mutations of BCORL1 in blast cells (bottom panels) but not in their matched normal buccal mucosa (top panels). (B) Schematic of somatic mutations identified in the BCORL1 gene in primary AML cells and AML cell lines. Red arrows indicate truncating mutations; and black arrows, missense mutations. NLS indicates nuclear localization signal; LXXLL, Leu-Xaa-Xaa-Leu-Leu motif; ANK, tandem ankyrin repeats; aa, amino acids; and CtBP, C-terminal binding protein.
We also measured normalized mRNA levels for BCORL1 in cDNA made from RNA isolated from FACS-sorted AML blasts in an expanded cohort of AML cases. In summary, we found relatively uniform expression of BCORL1 and all studied cases expressed the gene, including those with BCORL1 mutations (supplemental Table 9).
Analysis of the BCORL1 gene status in the AML cohort using SNP 6.0 array profiling uncovered no microdeletions or gains spanning the gene. Furthermore, copy-neutral loss of heterozygosity was not identified (supplemental Figure 1).
DNMT3A mutations have recently been reported in 22.1% (62 of 281) of de novo AML5 and in 20.5% of predominantly new diagnosed AML.6 Similarly, somatic mutations in DNMT3A were identified in 23.2% (22 of 95) of the cases we analyzed (supplemental Table 10). The novel DNMT3A-related finding in our study was a frameshift mutation in normal buccal mucosa and in remission marrow and neoplastic cells of AML patient89 (supplemental Figure 2). No additional mutations of DNMT3A were identified in the patient's tumor; the patient had no other tumors and was 78 years old. These data raise the possibility that germline DNMT3A mutations can weakly predispose persons to the development of AML but not to other tumors. There was no history of AML in this patient's family, and the parents were unavailable to determine whether the mutation was de novo.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Leukemia & Lymphoma Society (Clinical Scholar Award, S.N.M.), Virginia and D. K. Ludwig Fund for Cancer Research, and National Institutes of Health (grants CA46592, CA43460, and CA57345).
National Institutes of Health
Authorship
Contribution: M.L., B.V., and S.N.M. conceived the study and supervised the work; M.L., P.O., Y.J., S.N.M., N.P., K.W.K., and B.V. performed the laboratory research and genomic data analysis; R.C., H.E., D.B., and S.N.M. enrolled patients and contributed and analyzed clinical data; and M.L., B.V., and S.N.M. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nickolas Papadopoulos, Ludwig Center for Cancer Genetics and Therapeutics and Howard Hughes Medical Institute, Johns Hopkins Kimmel Cancer Center, 401 N Broadway, Baltimore, MD 21231; e-mail: npapado1@jhem.jhmi.edu; and Sami N. Malek, Department of Internal Medicine, Division of Hematology and Oncology, University of Michigan, 4312 Cancer Center, 1500 E Medical Center Dr, Ann Arbor, MI 48109; e-mail: smalek@med.umich.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal