Abstract
Abstract 4188
We recently demonstrated that a delayed time to find a compatible donor and also to proceed to allogeneic-HSCT can worsen the disease outcome (Michallet et al. ASH 2010). When a patient is presenting at our hospital for unrelated allogeneic-HSCT a donor search on the Bone Marrow Donor Worldwide (BMDW) registry is done, this query is limited to only 2-digits HLA typing of -A, -B, -DRB1 or -DQB1 loci. The identification of a compatible unrelated donor can be very long and expensive because of the limited HLA typing information in the registries. Tiercy et al. developed a computerized software that predicts the chance to find a suitable donor (BMT 2007, 40; 515–522) and the French transplant registry has recently developed a software called “Easy Match” that predicts the number of compatible donors for a given patient.
To refine and accelerate the process of donor search by combining the results of Tiercy score and EasyMatch applied on patients already received allogeneic-HSCT and define a new score for donor finding probability, in order to be time- and cost-efficient.
We retrospectively analyzed 104 adult and 34 pediatric patients who underwent allogeneic-HSCT between 2009 and 2011 after finding an unrelated donor (identical or not) or cord blood units. Firstly, we analyzed the HLA characteristics of each patient as previously described by Tiercy et al. to provide a HLA score with low, intermediate or high probability to identify a suitable identical donor.
Then, we used the EasyMatch software which realize a “qualitative” analysis that consist on checking that each HLA recipient phenotype was found among all possible pair wise combinations of 2 haplotypes of the different sets of haplotypes. Various “quantitative” analyses calculated the likelihood associated to each recipient phenotype for a given set of HLA genes, in a given population, at low versus high resolution typing. The EasyMatch software gives for each patient a number of potential donors sharing the same phenotype as the patient.
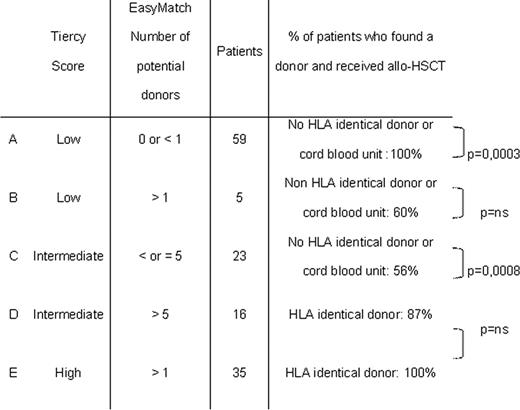
Our 138 patients were classified in 5 different categories (A to E) according to the combined results of the HLA score (Tiercy) and the EasyMatch software (Table).
The results of the combination of the two methods associated to the characteristics of our population, allowed the definition of a new scoring system applicable to each patient.
(group A): 0% of chances to identify a 10/10 identical donor for the recipient. The choice of the source will be defined considering the HLA characteristics of the recipient; in case of class I rare allele or rare HLA-BC linkage disequilibrium; a cord blood unit will be easier and more rapidly available. A complementary help should be given by an associated analysis with 4-digit haplotypes as defined by Maiers (Human Immunology 2007, 68; 779–788).
(groups B and C): about 50% of chances to identify a suitable 10/10 identical donor.
(groups D and E): from 75 to 100% of chances to identify a suitable 10/10 identical donor.
In conclusion, the use of this new scoring system allows time and cost spare. In case of low chance to find a donor, physicians can have a fast redirection to find another treatment alternative in order to maintain optimal results.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal