Abstract
Abstract 3853
According to the WHO-2008 classification, atypical chronic myeloid leukemia (aCML) is a myelodysplastic/myeloproliferative disorder clinically resembling the Philadelphia positive CML but lacking the BCR-ABL fusion. In aCML, the molecular lesions underlying the onset of the disease are unknown. To investigate the somatic events occurring in the genome of the aCML leukemic cells, we carried out whole-exome high-throughput sequencing analyses of 8 aCML patients. The results are described here.
Peripheral blood (PB) or bone marrow cells were obtained after informed consent at diagnosis, before any therapy. Myeloid cells, evaluated by FACS, constituted more than 80% of total cells. Lymphocytes were obtained from PB samples, after culture with PHA/IL2 for 2–3 weeks. The exon-capture protocol was performed on myeloid leukemic cells and normal lymphocytes from the same patients using the Illumina TruSeq Exome Enrichment Kit. The enriched DNA was sequenced with a Genome Analyzer IIx (Illumina), using a 60 bases paired-end protocol and the TruSeq chemistry. On average, 10.5 Gigabases per exome were generated. The bioinformatic analysis was performed using the Galaxy framework (http://main.g2.bx.psu.edu/); the cross-match between leukemic and normal exomes was performed with dedicated in-house C# software.
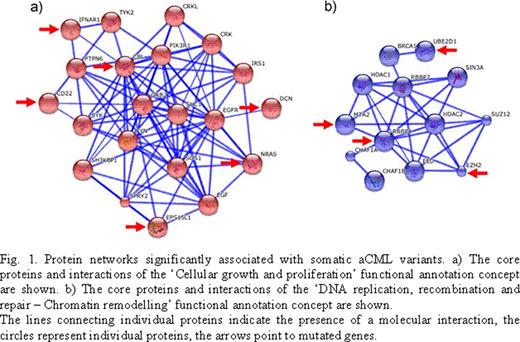
The percentage of reads matching the reference human genome was over 90%, with a mean exon coverage of >70-fold and a percentage of exons with a mean coverage ≥ 20x of > 90% for both the leukemic sample and the control. The percentage of nucleotides targeting exonic regions or exonic regions plus 100bp was 48% and 68%, respectively, with an overall 28-fold enrichment for exonic vs. non-exonic regions. The comparison between the leukemic and the control datasets led to the identification of 63 single nucleotide somatic mutations with a relative mutation coverage of > 35%, corresponding to their heterozygous presence in >88% of cells. In total, 46 mutations were transitions and 17 transversions (transition/transversion ratio of 2.7), with the C:G->T:A event occurring at highest frequency (35/63; 55.6%). In 19/35 (54.3%) of the cases, the C:G->T:A transition occurred in the context of a CpG site. Among the 63 variants, 32.8% ranked more than 1.0 and 21.3% more than 2.0 in the GeneRanker cancer scoring system (http://cbio.mskcc.org/tcga-generanker/). Characterization of the top scoring biological functions (Ingenuity Pathway Analysis software) revealed a strong association with the core functional concepts of ‘Cancer/Leukaemia' (p = 3.74 * 10−6) and ‘Myeloproliferative disorder' (p = 9.91 * 10−6), with a total of 23 genes in the Cancer and 18 in the Leukaemia functional annotation. The top scoring cellular functions were connected with the core concepts of ‘Cellular growth and proliferation' (p = 1.03 * 10−3; 17 genes) and ‘DNA replication, recombination and repair' (5 genes). The latter comprised the functional annotations: ‘Chromatin formation', ‘Chromatin remodelling' and ‘Formation of chromosome components' (p = 6.76 * 10−3, 1.96 * 10−2 and 7.76 * 10−4, respectively). A Markov Cluster Algorithm analysis (String, http://string-db.org), confirmed the presence of the same two main mutational clusters (Fig. 1a,b). The individual mutations identified will be presented at the meeting, including two recurrent mutations. Taken globally, these data suggest that somatic mutations targeting a specific cellular proliferation pathway and a chromatin remodelling protein network (Fig. 1a,b) may play a critical role in the onset/clonal evolution of aCML. This information may prove useful in the next future in order to develop evidence-based aCML treatment protocols.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal