Abstract
Abstract 3695
The heterogeneity of diffuse large B-cell lymphoma (DLBCL) has prompted the search for new markers that can accurately separate prognostic risk groups. We previously reported in multivariate analysis that Skp2 high expression by immunohistochemical method was a strong predictor of poor outcome in DLBCL patient treatment with CHOP and CHOP-R. (Ann of Oncology, 2010 21:833). However the precise mechanisms that regulate Skp2 expression in lymphoma are not fully understood. To better characterize the molecular mechanisms, we performed the DNA microarray study on uniformly treated DLBCL patients.
Total RNA was extracted from fresh tumor samples. All patients were diagnosis of de novo DLBCL. We compared with high(n=12) and low-Skp2(n=10) groups. Gene expression profiling was performed with the use of Illumina Human WG-6 array. The raw signal intensities of all samples were log2-transformed and normalized by rsn algorithm with □elumi' library package on Bioconductor software. We selected the probes, excluding the control probes, where the detection p-values of all samples were less than 0.01 and use them to identify differentially expressed genes.Then we applied Linear Models for Microarray Analysis (limma) package of Bioconductor software. To identify up or down-regulated genes between high and low Skp2, we calculated Z-scores and ratios (non-log scaled fold-change) from the normalized signal intensities of each probe.
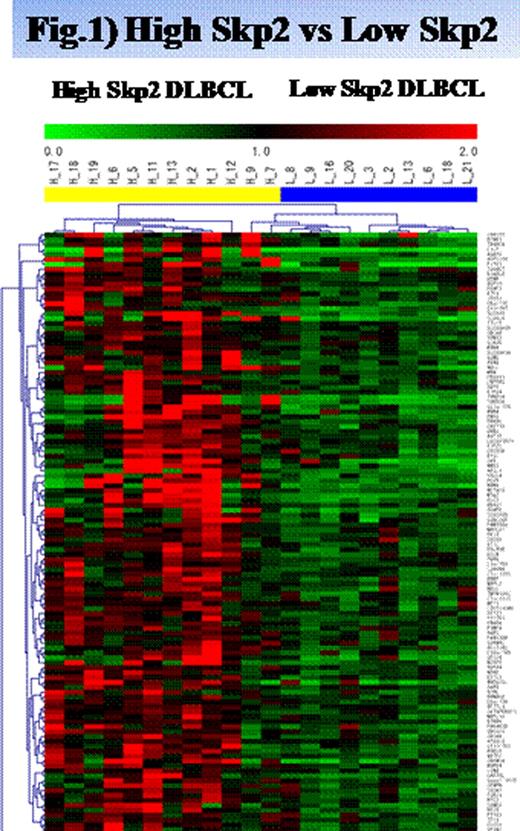
We selected 633 genes, 311 upregulated and 322 downregulated, which showed significant differences with a P-value of <0.01. Heat map of the differentally expressed genes between high and low-Skp2 groups (Figure 1). In addition, IPA (Ingenuity pathway analysis) showed clearly the network composed of cell cycle regulators, which mediate cell cycle progression during the G1/S checkpoint. A computer-assisted approach was used to procure specific molecular signalling pathways that were aberrantly expressed in DLBCL cells. Several genes related to cyclins and cell cycle regulation and to the MAPK, WNT, NF-kƒÀ and Myc mediated apoptosis signalling pathways were altered in DLBCL cells when compared with Skp2 levels. Gene highly expressed in high Skp2 included CDKN2A, MTA3, and TUBB2A. CDKN2A is known as a gene encoding a CDK inhibitor which functions in cell cycle regulator and the regulator of Myc pathway, and TUBB2A is known as a regulatory gene of the Mitosis. Recent studies have revealed that Skp2 is a transcriptional coactivator of Myc. MTA3 (Metastasis –associated protein 3), which was direct corepressor WNT4 pathway, was a high average increase (1.16-limma/log-fold change, respectively) in high Skp2 DLBCL. Although the basis for differentiation blockade is unkown in DLBCL, recent data suggest that BCL6 binding to the MTA3 corepressormight be involved. Additional biochemicaland functional studies are needed to determine whether MTA3 is a transcriptional factor of the Skp2 oncogene. The synergistic interaction of Myc, MTA3, and Skp2 may enhance the aggressiveness of Skp2-overexpressing lymphoma cells.
These genes may play a significant role in the pathogenesis of DLBCL anddeserve further investigation as candidates for new therapeutic targets.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal