Abstract
Abstract 3687
Multiple gene expression-based biomarkers have been identified in diffuse large B-cell lymphoma (DLBCL) that are predictive for survival outcomes. Most studies assess predictive significance based on p-value from multivariate Cox regression; few investigations have evaluated the incremental usefulness of these biomarkers in risk prediction. Using the recently developed concordance measures (e.g., C-statistics) on censored survival data, we assessed the usefulness of two published gene-based risk signatures and compared them to the known clinical prognostic factors; with an overall goal of investigating the added value.
The added value of biomarkers was assessed by C-statistic and the Integrated Discrimination Improvement (IDI). The overall C-statistic is an estimated concordance between prediction and observation (event vs. non-event) - the probability that predicted risk score is higher for subject with earlier time of event. The IDI measures overall improvement in sensitivity and specificity. The signatures we selected are a six-gene predictor by Lossos et al. (2004) and a three-component signature (∼400 genes) by Lenz et al. (2008). We used the Lenz dataset which include 233 patients with DLBCL who received R-CHOP therapy (median follow-up=2.81 yr), and focused on predicting 3-year survival outcome (42% censored). Clinical prognostic factors evaluated are the traditional IPI components (stage, performance status, age, LDH, and number of extra nodal sites).
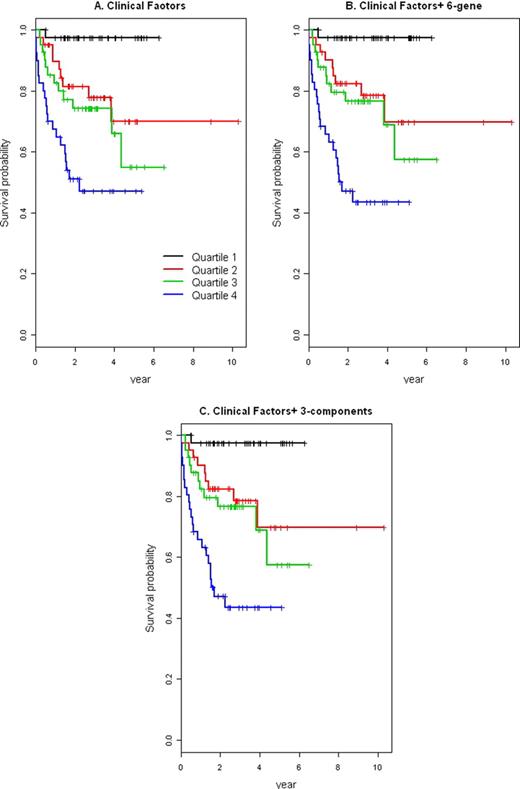
The C-statistic was 0.60 and 0.721 for six-gene predictor and three-component signature, suggesting good discrimination ability by molecular signature when used alone. However, the performance is inferior to IPI risk factors, with a C-statistic of 0.733. When integrating gene signatures with IPI risk factors, the C-statistic was increased to 0.744 and 0.762, an improvement of only 0.011 (95% CI, -0.049 to 0.071) and 0.029 (95% CI, -0.033 to 0.091) for six-gene predictor and three-component signature, respectively. Furthermore, assessment by IDI reveals an added value of only 0.011 (95% CI, -0.008 to 0.081) and 0.076 (95% CI, 0.013 to 0.16) for the two molecular signatures. Kaplan-Meier survival curves for the four quartile groups based on the predictor scores confirms the marginal benefit in risk prediction using molecular signatures. (Figure 1).
Overall survival for four quartile groups using clinical factors only (left), clinical factors+ 6-gene predictor (middle) and clinical factors+ 3-components signature (right).
Overall survival for four quartile groups using clinical factors only (left), clinical factors+ 6-gene predictor (middle) and clinical factors+ 3-components signature (right).
These results indicate that molecular biomarkers are inferior to clinical factors for risk assessment in DLBCL and provide little added value in risk prediction. These calculations suggest we will need to consider more than gene expression to develop highly discriminatory risk prediction models. However, the study of gene expression and clinical outcomes retains considerable potential to enhance understanding of disease mechanisms and uncover new therapeutic targets.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal