Abstract
Abstract 3561
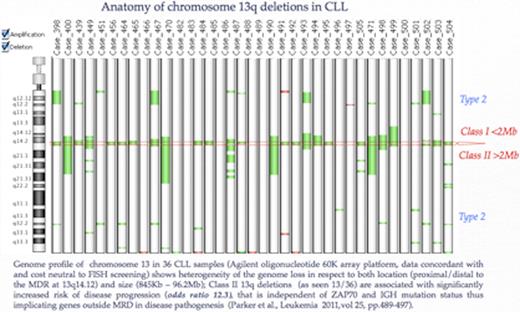
Specific genome aberrations are recognised prognostic factors in CLL. FISH-based genome risk classifications used in clinical decision making for over a decade. However molecular karyotyping is gaining acceptance as an alternative, albeit cost and skill demanding, that not only overcomes the limitations of FISH but provides comprehensive whole genome scanning. Indeed, several recent studies (Rinaldi et al., BJH 2011, Parker et al., Leukemia 2011, Quilette et al, Blood 2011) using different array platforms have revealed novel clinically relevant cryptic genome aberrations. Here we compare FISH and molecular karyotyping data of 40 diagnostic and follow up samples from 22 CLL patients. The whole genome screening (WGS) was performed with 8×60K oligonucleotide arrays (Agilent) using DNA from mononuclear fraction of p.blood/bone marrow samples as well as from fixed chromosome preparations and commercial reference DNA. Following manufacturerÕs protocols and bio-informatics routines (Z score and ADM1/2 algorithms) WGS was done at a cost comparable to the expense of routine FISH screening for 13q14, p53, ATM loss and trisomy 12. Genome array screening identified clonal imbalances recognised by FISH if present above 13%. In addition, WGS provided novel information in 73% of the samples, the results of which are summarised as follows (see figure below): (i) The 13q genome loss found in 10/13 presentation samples was stratified by size and location resulting in a positive assessment for 40% of the cases positive by FISH for del 13q14 as class II/type 2 deletions. These are known to be associated with high risk for disease progression; (ii) A common deleted region of 206.2Kb at @ 50556688–50699677 (hg19) was found to encompass the DLEU2 gene and is seen in 5/13 presentation samples with 13q loss; (iii) In 3 out of 13 cases FISH-positive for 13q loss, disease progression was accompanied by a proximal extension of the deletion to include the RB1 gene region thus acquiring the status of class II/type 2 deletion (high risk) (iv) Genome loss involving IGH sequences at 14q32.32, consistent with B cell type VDJ gene rearrangements was detected in all samples, while deletions within IG kappa and IG lambda were found in 5/25 patients; (v) Cryptic deletion of the ATM gene region was detected in 2 samples undetected by FISH; (vi) Total genome imbalances (TGA) in presentation samples varied between 3 and 17 with higher levels found in samples with p53 loss and class II, type 2 13q loss; (vii) Persistent disease was found to be associated with increase of TGA, some of which appeared to be non-random, such as loss of the whole or part of chromosome 19 and deletions of 4q21 region.
In conclusion, the application of 8×60K genome arrays provides established and growing genome-wide clinically relevant information as a cost neutral assay. A strategy by which array interrogation is carried out at presentation, followed by therapy response monitoring using FISH on a selected marker in CLL is a plausible alternative to routine practice. In cases where FISH has provided evidence for persistent disease, array screening will assess the genome damage, especially if carried on CD19(+)cell isolates, to assist treatment decisions.
Disclosures:
No relevant conflicts of interest to declare.
Author notes
*
Asterisk with author names denotes non-ASH members.
© 2011 by The American Society of Hematology
2011

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal