Abstract
Abstract 3447
Several studies have focused on the identification of biological markers that influence the clinical heterogeneity of Chronic lymphocytic leukemia (CLL). Until now IGVH and ZAP-70 are the most relevant prognostic markers in CLL suggesting the separation of two patient subgroups: with good MTZAP-70− (mutated and ZAP-70 negative) and poor UMZAP-70+ (unmutated and ZAP-70 positive) prognosis.
Gene expression profiling in CLL has been demonstrated to be useful for the development of new biomarkers as prognostic factors able to predict the clinical course, the response and resistance to therapy.
The aim of this study has focused on the identification of different molecular signatures capable of revealing new potential molecular predictors for prognostic assessment or genetic risk and intriguing biological pathways.
We have determined gene expression profiling of B cells in 112 CLL patients divided into three classes: the first with MT and ZAP-70−, the second with UM and ZAP-70+, and the third included both UM ZAP-70− and MT ZAP-70+ using Affymetrix HG-U133 Plus 2.0.
Among the set of 65 genes identified by microarray analysis we have found: AGPAT2, APP, ARSD, CHPT1, CRY1, DCLK2, LPL, MBOAT1, P2RX1, RIMKLB, ZAP-70, ZNF66 whose expressions were higher in second class (UMZAP-70+) while ADAM29, EGR3, FGL2, FUT8, NRIP1, TGFBR3, YPEL1 showed higher expression levels in first class (MTZAP-70−). Both analysis of differential expression and cluster analysis revealed that CLL patients were better partitioned in two rather than in three classes, based on their expression profiles.
We have also noted that the expression signature differentiating between MTZAP-70− and UMZAP-70+ B cells from CLL patients showed that gene coding for enzymes which regulate lipid metabolism (sphingolipid/glycerolipid/glycerophospholipid) ARSD, LPL, MBOAT1, CHPT1, AGPAT4, AGPAT2, PLD1 were overexpressed in UMZAP-70+ compared to MTZAP-70−.
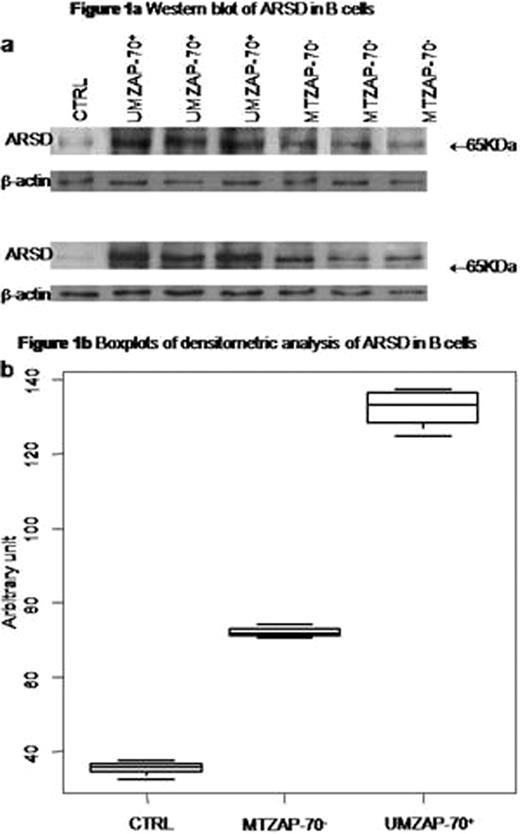
In particular we focused on ARSD as a new gene in CLL whose expression was consistently higher in UMZAP-70+ compared to MTZAP-70− CLL B cells.
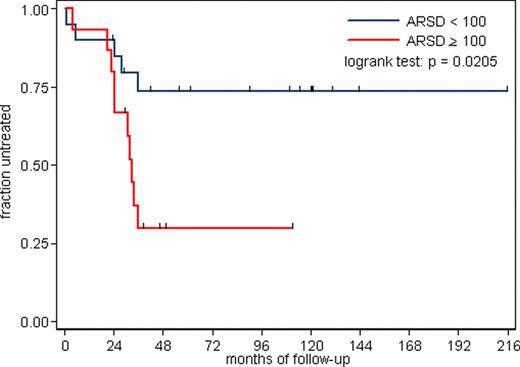
In conclusion 65 genes were differentially expressed in MTZAP-70− vs UMZAP-70+ CLL patients; among the 65 genes 7 were involved in lipid metabolism, in particular ARSD being identified in sphingolipid pathway. Western blotting experiments detected significantly higher ARSD levels in UMZAP-70+ compared to MTZAP-70− and CTRLs B cells.
Future studies will assess the role of ARSD as a prognostic factor in clinical trials combined with IGVH and ZAP-70 as well as the sphingolipid metabolism as a putative new biological mechanism in CLL.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal