Abstract
Abstract 3193
The switch from embryonic to fetal hemoglobin (HbF, α2γ2) in utero, and from fetal to adult hemoglobin at birth is well documented and achieved by the sequential activation of ε, γ and δ/β genes at the β globin gene (HBB) cluster. A change in the expression of hemoglobin genes also take place in adult erythropoiesis: earlier erythroid progenitors have been shown to produce significant amounts of fetal hemoglobin, while the more mature progenitors contain essentially none. In keeping with the sequential activation of β-like globin genes, δ globin chain synthesis also declines as maturation in erythroid progenitors progresses. Understanding the developmental changes of gene expression at the beta globin locus is not purely of academic interest, since a therapeutic induction of HbF or HbA2 (α2δ2) production would be of significant clinical benefit for patients with a defect of HbA (α2β2) function or abundance, such as sickle cell disease or β thalassemia. We have previously studied the genetic regulation of fetal hemoglobin persistence in a genome-wide association study (GWAS) in healthy volunteers, and are now extending this approach to the study of HbA2.
Our study population is the 'Twins UK' twin registry of healthy Europeans, mostly female adult individuals, with genome-wide single polymorphisms (SNP) data and hemoglobin phenotypes for a primary study group (n=2,340) and a second replication group (n=1,880). A quantitative trait GWAS analysis was carried out to assess the relationship between SNPs and the HbA2 trait.
We found that HbA2 (as a percentage of total hemoglobin) was weakly, but significantly, correlated with the amount of fetal hemoglobin carrying cells (F cells) an individual possesses (r = 0.14, p < 0.01). This suggests the existence of some common biological process that influences both hemoglobin species. We also found that the same SNP alleles at chromosome 6q23.3 (HBS1L-MYB, peak signal rs7775698, p = 2.51×10−9) that are associated with a boost in the prevalence of F cells and larger red blood cells (denoted by the mean cell volume or MCV) also promote HbA2 levels, again pointing to some common biological factor connected with the erythropoietic maturation process.
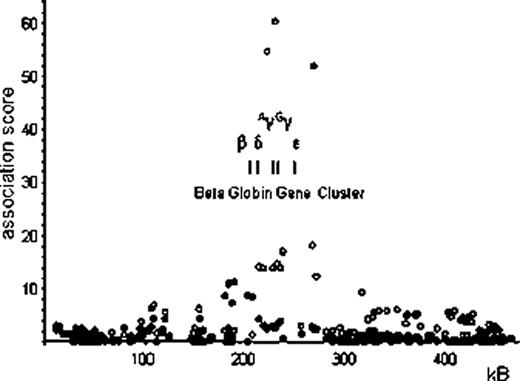
Interestingly, neither of the other two major HbF loci, BCL11A on chromosome 2p, or the HbF-promoting regions within the HBB cluster (at the β LCR and the γ globin genes) on chromosome 11p, showed association with HbA2 levels. Instead, SNPs around the β globin gene itself (clearly separate also from the delta gene) exert a significant influence on HbA2 levels (peak association rs12793110, p=5.11×10−12) (see Figure 1). In contrast to the HBS1L-MYB region on chromosome 6, the HbA2-boosting alleles at these SNPs do not increase red blood cell MCV. We propose that the SNPs around HBB influences HbA2 (ie. δ globin gene) expression via a mechanism that is related to the competitive process between the β and δ gene expression that might mimic a very mild β thalassemic effect.
Association with single-nucleotide polymorphisms (SNP) near the beta globin gene cluster on chromosome 11p15.4 with abundance of HbA2 (filled circles ¥) and F cells (empty circles ○) in the peripheral blood of Northern European adults (Twins UK).
Association with single-nucleotide polymorphisms (SNP) near the beta globin gene cluster on chromosome 11p15.4 with abundance of HbA2 (filled circles ¥) and F cells (empty circles ○) in the peripheral blood of Northern European adults (Twins UK).
We propose that the systematic genetic study of specialized hematological traits in healthy volunteers can help to understand the biology of hematopoiesis.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal