Abstract
Abstract 2246
Factor IX (FIX) is a vitamin K-dependent protein that circulates in blood as a zymogen of the serine protease factor IXa (FIXa). During physiologic coagulation, it is activated by FXIa/Ca2+ and FVIIa/tissue factor/Ca2+. Activation of FIX by either enzyme occurs in two sequential steps to produce an active serine protease and release of a 35 residue activation peptide of amino acid residues 146–180. FIX can also be activated by a protease from Russell's viper venom (RVV). The N-terminal light chain of FIXa contains the g-carboxyglutamic acid domain followed by two epidermal growth factor-like (EGF1 and EGF2) domains, whereas the C-terminal heavy chain contains the serine protease domain. FIXa activates FX requiring Ca2+, phospolipid, and FVIIIa. Previously, we reported that protease domain of FIXa contains a Ca2+-site similar to that in trypsin and a Na+-site similar to that in FXa and activated protein C (APC). These Ca2+- and Na+-sites play an important role in FIXa physiologic function (Schmidt et al, J Mol Biol, 350, 78–91, 2005). Subsequently, three studies raised concerns regarding the existence of Na+-site in the protease domain of FIXa. One of the studies is biochemical in nature (Gopalakrishna and Rezaie, Thromb Haemost, 95, 936–941, 2006) and the other two are structural studies with either mutations in the protease domain of FIXa (Zogg and Brandstetter, Structure, 17, 1669–1678, 2009) or with the protease domain bound to antithrombin (Johnson et al., PNAS 107, 645–650, 2010). To address these concerns, we obtained EGF2/Protease domain of wild type FIXa and determined its structure in the absence of any macromolecular ligand. The EGF2/Protease domain in the zymogen form was expressed in E. Coli, and purified using FPLC gel filtration and ion exchange chromatography. The isolated protein was activated with RVV and the EGF2/protease domain was further purified using FPLC HiTrap heparin and Benzamidine column chromatography. The purified EGF2/Protease domain was fully activated as analyzed by reduced SDS-PAGE. The protein in 1 M NaCl/Tris buffer was crystallized using PEG 4000 and 0.2 M ammonium sulfate as precipitants. One of the crystals diffracted to 1.64 Å, and belonged to the space group I4122 with cell dimensions a=b=110.68 Å, c=113.38 Å. The data were processed with XDS and the structure was solved by molecular replacement using 1RFN as the starting model. Refinement was carried out using Refmac from the CCP4 package. In our structure, Na+-site in the protease domain is clearly defined. It involves coordination from the four predicted main chain carbonyl groups and two water molecules in the octahedral geometry (Fig. 1). The structure is presently refined with Rwork and Rfree of 21.7 and 25.6, respectively; further refinement is in progress.
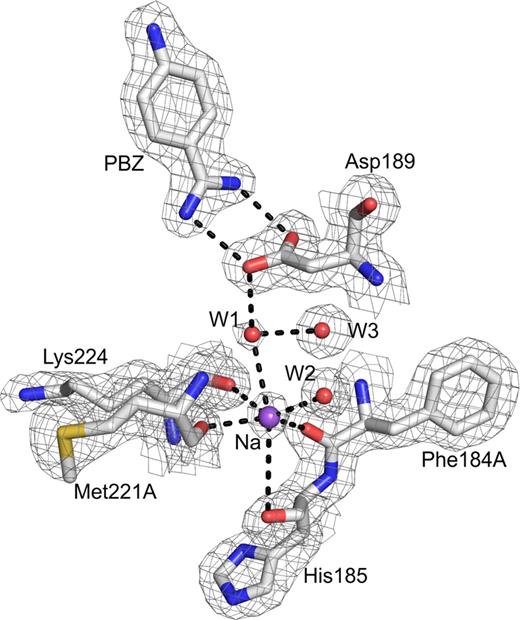
Na+-site in FIXa protease domain. The Na+-site in FIXa involves coordination from four carbonyl groups (184A, 185, 221A and 224) from the protein and two water molecules, W1 and W2. Electron density (2Fobs- Fcalc) is contoured at 1σ and sodium is labeled as Na. Note that the Na+-site is linked through water to the carboxylate of Asp189. The salt bridge between PBZ and Asp189 at the S1 site is shown. The coordination of the Na+-site is similar to that in FXa, FVIIa and APC. It differs from thrombin where two main chain carbonyls and four water molecules are coordinated to Na+.
Na+-site in FIXa protease domain. The Na+-site in FIXa involves coordination from four carbonyl groups (184A, 185, 221A and 224) from the protein and two water molecules, W1 and W2. Electron density (2Fobs- Fcalc) is contoured at 1σ and sodium is labeled as Na. Note that the Na+-site is linked through water to the carboxylate of Asp189. The salt bridge between PBZ and Asp189 at the S1 site is shown. The coordination of the Na+-site is similar to that in FXa, FVIIa and APC. It differs from thrombin where two main chain carbonyls and four water molecules are coordinated to Na+.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal