Abstract
Abstract 1447
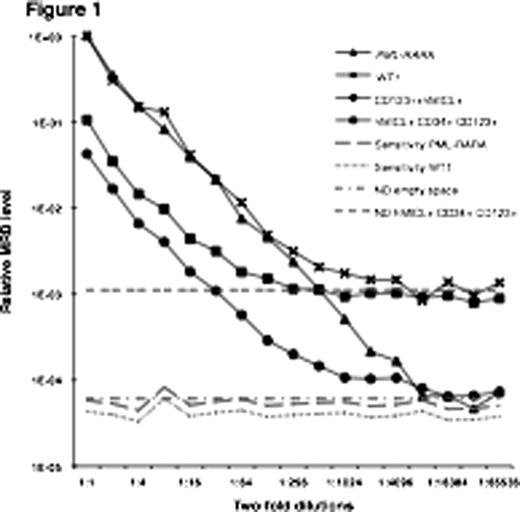
There is a growing realization that outcome of acute myeloid leukemia (AML) is strongly correlated to whether minimal residual disease (MRD) - as quantified by either multicolor flow cytometry (MFC) or real-time quantitative polymerase chain reaction (qPCR) - is demonstrable at crucial clinical decision time points. While FCM is applicable in up to 95% of all cases through the detection of leukemia-associated immunophenotypes (LAIPs), its major drawback is the lack of sensitivity in most cases and antigen shift, which is seen in up to 20% of cases. In contrast, while its sensitivity is generally considered to be higher than that of FCM, qPCR can only be applied in less than 80% of the patients. We have previously reported human Myeloid Inhibitory C-type Lectin (hMICL) and the interleukin-3 receptor alpha (CD123) to be potent and widely applicable markers in MRD detection by MFC, but the sensitivity of this approach compared to qPCR techniques remains to be elucidated (Clinical Cytometry, Part B, in press). Problem formulation: No previous studies have to our knowledge established the sensitivity levels of markers within these methodologies. Data from such experimentation would allow for better decision making as to what marker to apply in the given patient. Hypothesis: We hypothesized that the direct comparison between MRD technologies might yield hard data as to optimal choice of methods in the pre-clinical setting, and ultimately also for design of future FCM based MRD protocols. Results: Initially, we determined the validity of hMICL/CD123 FCM assay by performing two-fold dilution series of cryopreserved AML cells in highly purified T-cells, the latter being hMICL-/CD123-/CD34-/CD117-. Dilutions at 1: 1 to 1: 214 displayed linearity in MRD drop and resulted in a coefficient of correlation of 0.99 (P = <0.01) for all experiments. The sensitivity was calculated from the plateau-phase at of the titration curve at 1: 216 and reached a maximal sensitivity of 1: 30.000 in this setting. In order to evaluate the sensitivity of the hMICL/CD123 technique compared to standard qPCR methods in experiments, we admixed leukemic blasts from 5 patients with either known fusion transcript positive and WT-1 + disease in normal bone marrow. Sensitivities were calculated from the plateau phase at 216. The most sensitive LAIP was defined in each patient from internal standard references generated from normal donors and regenerating BM from acute lymphoblastic leukemia patients. The detection limits of MFC ranged from 10−3 to below 10−4 for the most sensitive hMICL/CD123 combinations. While the fusion transcript qPCR assays were found to be the most sensitive, the hMICL/CD123 FCM one stood up surprisingly well in 3 of 5 cases of fusion transcript positive disease and approached the qPCR assay. In all 5 cases the FCM did better than the WT1-assay (an example of titration curves and relative MRD values for a PML-RARA + patient is given in the Figure). In the clinical setting, we analyzed a total of 69 unselected AML patients consecutively diagnosed at our department. A total of 10 different hMICL/CD123 LAIPs were analyzed closely, of which one or more were present in 90% of the patients. By calculating log differences between diagnostic AML samples and normal and regenerating BM we found the frequency of LAIP+ cells to be more than 4 logs higher in leukemia than in normal BM for the most sensitive combinations of LAIPs. Conclusion: These data are the first to directly compare MRD sensitivity of FCM and qPCR in spiking experiments and in a larger patient material. They reveal that a new assay involving hMICL and CD123, which are stable markers throughout the disease in these patients, has a surprisingly high sensitivity, though not quite at the level of standard qPCR assays. These data can be used for the design of future studies of cytoreduction in AML, where individualized therapy is warranted.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal