Abstract
Abstract 132
There is increasing evidence on the importance of epigenetic mechanisms such as DNA methylation and acetylation in the pathogenesis of multiple myeloma (MM). A global DNA hypomethylation pattern with selective genes hypermethylated has been described in myeloma plasma cells when compared with normal plasma cells. This fact could constitute a potential target for the use of demethylating agents. The response to bortezomib, a widely used agent against myeloma cells through proteasome inhibition, is particularly variable in patients with relapsed or refractory disease. We examined both, the global DNA methylation pattern and methylation state in 30 genes, in DNA from bone marrow cells and correlated our findings with response, progression (PFS) and overall-survival (OS) to bortezomib in patients with relapsed myeloma.
Seventy-five patients (37M/38F; median age 65 years, range 29 to 80) with relapsed MM were treated from December 2002 to March 2010 with bortezomib-based regimens at our institution. Median follow-up for patients alive was 31 months (range 6 to 45). Genomic DNA was isolated from bone marrow slides with plasma cell infiltration at the time of relapse using a commercial kit (Qiagen). Global methylation was determined in all patients by ELISA (Epigentek), obtaining the percentage of 5-methylcytosine (5-mC) present in total DNA. CpG island DNA methylation profile of 30 genes was determined in 42 patients by a DNA methylation PCR system based on methylation sensitive and/or dependent restriction enzymes digestion (Qiagen). These genes were selected based on either their potential impact on prognosis in previous reports, or on the pathogenesis of MM, involving several cellular pathways such as innate immune response (CD40, EP300, MIF, CBP, TGFB1, TGFBR2), cytokine receptors (CXCR4, CXCL12, IL6R, IL17RA), transcription factors (NFKB1, NFKBIB, IRF4), cytokine stimulus response (SOCS3), apoptosis (TNFRSF13C, TNFRSF21, TNFRSF25, BCL2L11), tumor suppression (TP53, BRCA1, DAPK1, CDH1, RASD1), cellular cycle control (CCNB1, CCND1, CCNA2, CCNE1, CDKN2A, CDKN1A) and efflux transporter (ABCG2).
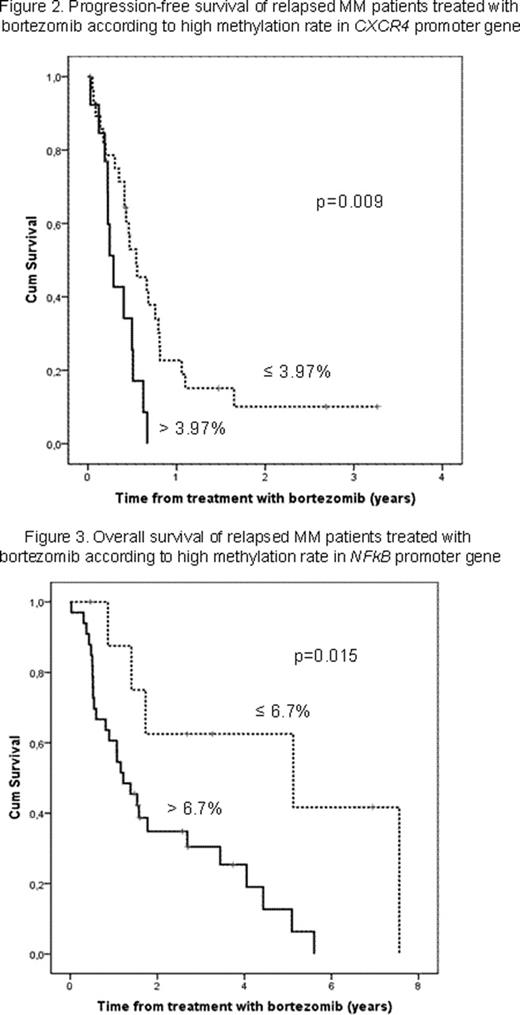
Overall response (OR) was achieved in 62% of the patients (complete remission 6.7%, partial response 44% and minor response 10.7%), while 9 (12%) and 20 (26.7%) showed no response (NR) or progressive disease (PD), respectively. The median PFS and OS after bortezomib therapy were 6 and 19.6 months, respectively. A low global methylation status was observed (median 4.68% of 5-mC, range 0.02 to 13.6) and patients with more than 3.95% of total DNA methylated achieved better OS than patients with more unmethylated DNA (median 30 versus 15 months) (p=0.004; Figure 1). Concerning methylation on specific-genes, a methylation status lower than 3.97% in CXCR4 was correlated with a longer PFS after bortezomib treatment (p=0.009; Figure 2). Clustering analysis with methylation status for these genes, showed that NFkB presented a differential profile according to response to bortezomib (p=0.037). A relative low methylation percentage (lower than 6.7%) in this gene was also associated with longer OS after bortezomib treatment (p=0.015; Figure 3). A positive correlation was observed with high methylation status in NFkB and other genes involved in the same cellular pathway (NFKBIB, EP300, CBP, CCNA2, CCNB1) (p<0.025). Moreover, a combination of highly methylated global genome and low NFkB methylation status defined a specific subset of patients with better prognosis (p=0.005) in terms of OS. Finally, a multivariate analysis including number of previous treatment lines, autologous stem-cell transplantation, previous exposure to bortezomib as well as global and NFkB methylation status showed that only the last two variables retained significance (p=0.035, OR=0.43 and p=0.028, OR=3.4, respectively).
In our study, a low methylation grade in the overall DNA was observed. A relative high methylation status in the global genome and low in NFkB were associated with longer OS after bortezomib therapy in patients with relapsed or refractory myeloma. These results could be explained through the potential cell effect mediated by bortezomib in the NFkb pathway. Finally, a subgroup of patients with an ominous prognosis associated with DNA methylation at relapse in spite of bortezomib treatment was identified.
Fernández de Larrea:Novartis: Honoraria; Janssen: Honoraria; Celgene: Honoraria. Cibeira:Janssen: Honoraria; Celgene: Honoraria. Rosiñol:Janssen: Honoraria; Celgene: Honoraria. Blade:Janssen: Honoraria; Celgene: Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal