Understanding the mechanism of platelet activation requires the knowledge of the structural organization of the glycoprotein (GP) Ib-IX-V receptor. In this issue of Blood, an innovative structural approach was used to shed light on the interaction between GPIbβ and GPIX subunits.1
The GPIb-IX-V complex serves as the platelet receptor that mediates binding to collagen-bound von Willebrand factor (VWF) at sites of vascular injury, leading to platelet activation and thrombus formation.2 GPIb-IX-V also binds many other ligands, the functional consequences of which are more obscure; in the case of thrombin even despite the availability of structural information.3 The process of platelet activation must be tightly regulated because dysregulated thrombus formation blocks blood vessels and leads to ischemia, for example, in heart attack and ischemic stroke. Defects in GPIb-IX-V lead to the bleeding disorder Bernard-Soulier syndrome.
Three-dimensional structural information of components of signaling pathways and their interactions is essential for an in-depth understanding of processes such as platelet activation. The key method for obtaining atomic resolution structural information is X-ray crystallography, a technique accounting for 88% of structures deposited in the Protein Data Bank. Unfortunately, this technique requires milligram amounts of purified protein, which has to subsequently be induced to form crystals. Both protein production and crystallization can be difficult tasks particularly for membrane-bound, multidomain and multisubunit macromolecular complexes. GPIb-IX-V demonstrates all these difficulty criteria. In such cases structural biologists use the divide-and-conquer approach, dividing the proteins into smaller, more manageable fragments, for example, deleting the membrane-spanning regions. These approaches previously led to structure determination of the extracellular domain of GPIbα and its complex with VWF-A1 domain.4,5 A strategy that can be used in even more stubborn cases is to crystallize the protein of interest in the presence of large fusion-tag, as recently exemplified for β-adrenergic and Toll-like receptors.6 This approach has been taken a step further by McEwan et al, who grafted 3 segments of the GPIX subunit onto the homologous GPIbβ subunit (see figure).7 McEwan et al now describe the structures of the extracellular domains of both GPIbβ and this GPIbβ/IX chimera.1 The observation that GPIX cannot be expressed on cell surface by itself, but only in the presence of GPIbβ, provides an assay for the effects of mutations on the interaction between these proteins through the analysis of surface expression in transiently transfected cells.
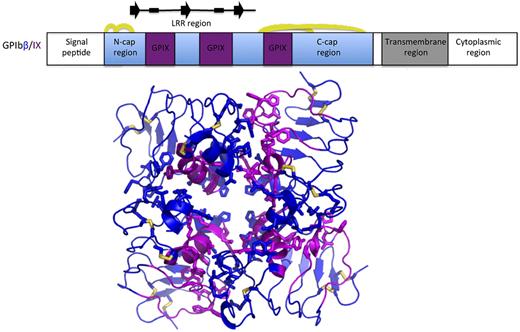
McEwan et al used an innovative chimeric protein approach to obtain structural information on the GPIbβ-GPIX association in the GPIb-IX-V complex.1 The top panel shows a schematic diagram of the sequence of GPIbβ, with the portion of GPIbβ used for structural analysis shown in blue. The GPIX sequences grafted into the GPIbβ scaffold in the GPIbβ/IX chimera are shown in magenta. Disulfide bridges are indicted by yellow arcs, and elements of secondary structure in the solenoid repeat region are indicated above the diagram (arrows for β-strands, boxes for 310 helices). The bottom panel shows 4 protein chains from the crystal of the GPIbβ (blue)/IX (magenta) chimera in cartoon representation, highlighting the side chains of interface residues and disulfide bridges (yellow) in stick representation.
McEwan et al used an innovative chimeric protein approach to obtain structural information on the GPIbβ-GPIX association in the GPIb-IX-V complex.1 The top panel shows a schematic diagram of the sequence of GPIbβ, with the portion of GPIbβ used for structural analysis shown in blue. The GPIX sequences grafted into the GPIbβ scaffold in the GPIbβ/IX chimera are shown in magenta. Disulfide bridges are indicted by yellow arcs, and elements of secondary structure in the solenoid repeat region are indicated above the diagram (arrows for β-strands, boxes for 310 helices). The bottom panel shows 4 protein chains from the crystal of the GPIbβ (blue)/IX (magenta) chimera in cartoon representation, highlighting the side chains of interface residues and disulfide bridges (yellow) in stick representation.
One of the key advances provided by McEwan and colleagues is the description of the quaternary association of GPIb-IX sub-complex (the GPV subunit is not essential for most functions of the receptor). Clearly, one needs to be cautious when interpreting the interfaces observed in the GPIbβ/IX chimera crystals as reflective of the functional interaction between GPIbβ and GPIX in the intact receptor complex. However, this conclusion is the simplest explanation of the available data and is supported by several pieces of evidence: (1) it was demonstrated, based on surface expression analysis and coimmunoprecipitation experiments, that the GPIX loops grafted in the GPIbβ/IX chimera are sufficient to mediate association with GPIbβ7 ; (2) the crystal structure of the GPIbβ/IX chimera reveals interactions of GPIX loops with GPIbβ sequences that that are consistent with association in the native complex; (3) analogous interactions are observed in at least 2 crystal forms of the GPIbβ/IX chimeric protein; and (4) mutagenesis of residues in the observed interface results in a loss of the ability of GPIbβ to support surface expression of GPIX in transiently transfected cells. Because interactions between transmembrane helices make a major contribution to the association of receptor subunits, the self-association of the GPIbβ/IX chimera observed in the crystals is not observed in solution. The nature of self-association of the current GPIbβ/IX chimera makes it unsuitable to study its interactions with GPIbβ directly, but this may be possible after grafting further GPIX segments into the GPIbβ scaffold. While the current data allow a possible arrangement for the binding of 2 GPIβ subunits to GPIX to be deduced, as presented by McEwan et al, further data are required to develop a unique model.
The paucity of structural data on the GPIb-IX complex has also meant that the molecular basis of a number of mutations leading to Bernard-Soulier syndrome has remained unclear. A combination of structural and surface expression analysis now provides an immediate explanation for 8 mutations in the GPIbβ extracellular domain and suggests different mechanisms leading to disease. While 6 of the mutations affect proper folding and secretion of GPIbβ, 2 exhibit no such effect. Instead, these 2 mutations result in a lack of GPIX cell-surface expression when GPIbβ and GPIX are coexpressed, suggesting they perturb the functional interaction between GP1bβ and GPIX. Indeed, the corresponding residues are present near the GPIbβ-GPIX interface revealed by the GPIbβ/IX chimera structure.
The structures presented by McEwan et al also have general implications in terms of protein structure.1 GPIbβ and GPIX unusually contain only 1 copy of the leucine-rich repeat (LRR) structural motif. As the stability of LRR8 and more generally solenoid9 protein structures depends on stacking of several repeats, a single repeat would not be expected to adopt a stable structure. The structures reveal that a stable structure is formed by the stacking of 3 solenoid layers through the help of repeat-flanking sequences.
The new structural information not only advances the functional understanding of the GPIb-IX-V complex, but also provides new opportunities for developing antithrombotic therapeutics with a potential for reduced bleeding side effects.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal