In this issue of Blood, Wong and colleagues provide compelling evidence that Shwachman-Diamond syndrome is a ribosomopathy.1 Using both lymphoblasts from patient samples and an elegant model of engineered conditional mutants in Dictyostelium dicoideum, an amoebozoan, the authors provide compelling evidence that defective maturation of the 60S ribosomal subunit is fundamental to the pathophysiology of the disorder.
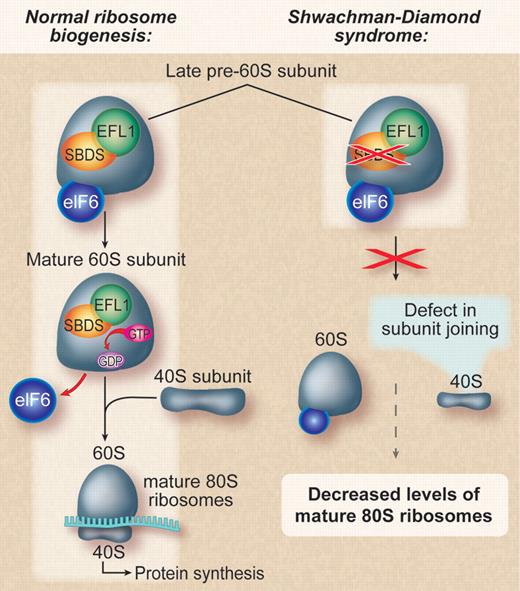
Proposed mechanism for the cellular consequences of mutations in Shwachman-Bodian-Diamond syndrome. Left panel: Normal cell with unperturbed ribosome biogenesis and proper joining of the ribosomal subunits. Right panel: Mutations in SBDS lead to a failure to evict eIF6 resulting in a defect in subunit joining and a subsequent decrease in levels of translationally active mature 80S ribosomes. Professional illustration by Debra T. Dartez.
Proposed mechanism for the cellular consequences of mutations in Shwachman-Bodian-Diamond syndrome. Left panel: Normal cell with unperturbed ribosome biogenesis and proper joining of the ribosomal subunits. Right panel: Mutations in SBDS lead to a failure to evict eIF6 resulting in a defect in subunit joining and a subsequent decrease in levels of translationally active mature 80S ribosomes. Professional illustration by Debra T. Dartez.
Shwachman-Diamond syndrome is a rare autosomal disease characterized by exocrine pancreatic insufficiency, ineffective hematopoiesis, and an increased risk for leukemia.2 In approximately 90% of patients, the disease is caused by biallelic mutations in the SBDS (Shwachman-Bodian-Diamond syndrome) gene.3 SBDS has been implicated in multiple biologic processes including ribosome biogenesis, stabilization of the mitotic spindle, and cell motility, but the functional defect that causes the Shwachman-Diamond syndrome phenotype has not been clear.4
Studies in yeast and mammalian cells have demonstrated a role for SBDS the final stages of 60S ribosome maturation and the joining of 60S and 40S ribosomes to form the translationally active 80S ribosome.5,6 Before nuclear export, premature joining of the ribosomal subunits is prevented by a protein called eIF6 (Tif6 in yeast) that binds the 60S ribosome at the intersubunit interface. After nuclear export, eIF6 is released, and the subunits join to form the mature 80S ribosome. In yeast, the SBDS ortholog (Sdo1) functions with ELF1, a GTPase, to release Tif6. The relevance of this model to the human disease has not been conclusively established as primary patient fibroblasts do not appear to have a defect in subunit joining.7
Here, Wong et al studied the sbds gene using an innovative model of genetically engineered amoeba, Dictyostelium dicoideum, a species that is distant from both yeast and mammals on the eukaryotic tree. The sbds gene is conserved in Dictyostelium, and conditional inactivation of the gene using temperature-sensitive, self-splicing inteins disrupts ribosome subunit joining. Remarkably, the human SBDS gene complements the mutant Dictyostelium. Variants of the SBDS gene with disease-causing mutations do not successfully rescue the mutants, demonstrating the deleterious effects of these mutations.
The authors went on to demonstrate that SBDS protein coimmunoprecipates with EFL1 and that the proteins share a physical proximity on the surface of the 60S subunit. Moreover, SBDS protein and EFL1 cooperate to cause the release of eIF6 in a process that requires GTP binding. The eviction of eIF6 allows proper joining of the ribosomal subunits and effective protein synthesis (see figure). These data corroborate another recent study using Sbds-deleted mice to show that SBDS protein and EFL1 directly catalyze the removal of eIF6, by a mechanism that requires GTP binding, allowing for the translational activation of ribosomes.6 Finally, Wong et al extended their work to human lymphoblasts from patients with Shwachman-Diamond syndrome. They found that SBDS protein expression was inversely correlated with the severity of ribosomal subunit joining defect.
Taken together, these data strongly indicate that Shwachman-Diamond syndrome is a ribosomopathy, adding another intriguing member to this fascinating class of diseases. Diamond Blackfan anemia (DBA), the first human disease to be linked to ribosome dysfunction, is characterized by a profound macrocytic anemia and a range of physical abnormalities including craniofacial and cardiac defects.2 Haploinsufficiency for 10 different ribosomal proteins has now been described in patients with Diamond Blackfan anemia, and acquired haploinsufficiency for RPS14 has been implicated in the 5q- syndrome, a subtype of myelodysplastic syndrome.2,8 The pathophysiology of these disorders involves the induction of p53, particularly in the erythroid lineage.9
Another disorder with convincing links to ribosomal dysfunction is Treacher Collins syndrome (TCS). Patients with Treacher Collins syndrome have craniofacial abnormalities that are similar to patients with Diamond Blackfan anemia, but do not develop bone marrow failure. TCOF1, the gene mutated in many patients with Treacher Collins syndrome, encodes a protein that is essential for the transcription of ribosomal DNA and may play a role in the methylation of rRNA.10 Moreover, mutations have recently been reported in Treacher Collins syndrome patients in genes encoding subunits of RNA polymerase I and III.11
A central unanswered question is how defects in ribosome biogenesis lead to divergent clinical phenotypes. Both Diamond Blackfan anemia and Shwachman-Diamond syndrome cause bone marrow failure, but patients with the former have a more severe defect in erythropoiesis, while the latter tend to have worse neutropenia. Patients with Treacher Collins syndrome and some with Diamond Blackfan anemia develop craniofacial abnormalities but patients with Treacher Collins syndrome have normal hematopoeisis. Developmental and tissue-specific gene expression or translational requirements may cause differential sensitivities to decreased expression of particular genes involved in ribosome function, but this remains to be elucidated.
The anemia in Diamond Blackfan anemia and the 5q- syndrome, as well as the craniofacial defects in Treacher Collins syndrome, appear to be caused by activation of p53 in distinct lineages. The degree to which p53 is pathologically activated in vivo by abnormal eIF6 release in Shwachman-Diamond syndrome remains to be determined. Despite the many unanswered questions, it is increasingly clear that genetic lesions causing specific defects in ribosome biogenesis are fundamental to the pathophysiology of multiple human disorders.
Conflict-of-interest disclosure: The authors declare no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal