Abstract
Recent studies have shown that 3-Deazaneplanocin A (DZNep), a histone methyltransferase inhibitor, disrupts polycomb-repressive complex 2 (PRC2), and preferentially induces apoptosis in cancer cells, including acute myeloid leukemia (AML). However, the underlying molecular mechanisms are not well understood. The present study demonstrates that DZNep induces robust apoptosis in AML cell lines, primary cells, and targets CD34+CD38− leukemia stem cell (LSC)–enriched subpopulations. Using RNA interference (RNAi), gene expression profiling, and ChIP, we identified that TXNIP, a major redox control molecule, plays a crucial role in DZNep-induced apoptosis. We show that disruption of PRC2, either by DZNep treatment or EZH2 knockdown, reactivates TXNIP, inhibits thioredoxin activity, and increases reactive oxygen species (ROS), leading to apoptosis. Furthermore, we show that TXNIP is down-regulated in AML and is a direct target of PRC2-mediated gene silencing. Consistent with the ROS accumulation on DZNep treatment, we also see a signature of endoplasmic reticulum (ER) stress-regulated genes, commonly associated with cell survival, down-regulated by DZNep. Taken together, we uncover a novel molecular mechanism of DZNep-mediated apoptosis and propose that EZH2 may be a potential new target for epigenetic treatment in AML.
Introduction
Acute myeloid leukemia (AML) is characterized by clonal expansion of immature malignant myeloid hematopoietic cells. Depending on the subtypes of leukemia and the underlying genetic defects, the long-term survival can range from 20%-90%.1 The treatment of leukemia has progressed little over the last decades and the mainstay is still chemotherapy.2 Therapy targeting epigenetic machinery is coming to the fore in cancers.3,4 Several studies have demonstrated the potential use of epigenetic treatment, such as vorinostat (suberoylanilide hydroxamic acid [SAHA]) and decitabine (5-aza-2′-deoxycytidine) for AML.5,6
Epigenetic modifications of chromatin structure achieved by the reversal of acetylation, methylation, and phosphorylation of core histone proteins in the nucelosomes play a pivotal role in normal HSC self-renewal, proliferation, and lineage commitment.7 Like its normal counterparts, epigenetic changes that perturb the balance of self-renewal and proliferation contribute to the transformation of leukemia stem cells (LSCs).8 Studies in Drosophila revealed that Trithorax group (TrxG) and Polycomb group (PcG) genes have opposite functions in the regulation of homeobox genes.9,10 This regulatory pathway appears to be evolutionarily conserved from Drosophila to human. Polycomb-repressive complex 2 (PRC2) mediates silencing of PcG-bound genes through trimethylation of histone H3 on Lys27 (H3K27me3).11 The core components of the human PRC2 are EZH2 (enhancer of zeste homolog 2), SUZ12 (suppressor of zeste 12), and EED (embryonic ectoderm development). EZH2 has a SET domain that harbors histone methyltransferase activity when complexed with SUZ12 and EED.12 Genome-wide studies show that the majority of PRC2 target genes are cell-fate transcription factors and signaling molecules regulating development and cell differentiation.13 In addition, the H3K27me3-enriched repressive chromatin structure is crucial for stem cells to maintain their quiescent and undifferentiated status.14 Overexpression of EZH2 has been identified in wide ranges of solid tumors including prostate, breast, colon, liver, bladder, as well as hematologic malignancies.14 Furthermore, high EZH2 expression has been associated with advanced stage, metastatic tumors, and poorer prognosis in cancers.15
We previously reported that 3-Deazaneplanocin A (DZNep), a S-adenosylhomocystein (AdoHcy) hydrolase inhibitor, depletes EZH2 and the associated H3K27me3, and induces apoptosis in breast and colon cancer cells.16 The effectiveness of DZNep has also been demonstrated by other investigators.17-22 In AML, DZNep treatment leads to apoptotic cell death in HL60, OCI-AML3 cell lines, and primary patient cells.22 Furthermore, we and others showed that DZNep is synergistic with histone deacetylase (HDAC) inhibitors and DNA methyltransferase (DNMT) inhibitors in activating silenced genes.19,22,23 In light of the important role in stem cell maintenance and tumor progression, we assess the effect of targeting EZH2 by DZNep treatment in human AML and LSC-enriched subpopulations. In this study, we showed that DZNep pharmacologically depletes PRC2 and decreases H3K27me3, resulting in up-regulation of thioredoxin-binding protein 2 (TXNIP). In turn, this caused an increase in reactive oxygen species (ROS) production. These studies dissect the mechanisms by which DZNep induces apoptosis in AML in vitro and in vivo and inhibits LSC.
Methods
Cell culture
A panel of acute myeloid leukemia cell lines, that is, MV4-11 (M5), MOLM-14 (M5), Mono-Mac-1 (M5, kindly provided by Dr Matiullah Khan, Cancer Science Institute, Singapore, National University of Singapore), THP-1 (M5), HL60 (M2), and KG-1 (M1) were cultured with RPMI 1640 (Invitrogen) supplemented with the addition of 10% FBS (JRH Bioscience Inc) at density of 2-10 × 105 cells/mL in a humid incubator with 5% CO2 at 37°C. TF-1 (M6) cells were cultured in the same medium with the addition of 5 ng/mL human IL-3 (PeproTech). Kasumi-1 (M2, kindly provided by Dr Motomi Osato, Cancer Science Institute, Singapore, National University of Singapore) cells were grown in RPMI 1640 (80%) with 20% FBS and 2mM L-Glutamine (Invitrogen). BM blast cells (> 90%) from newly diagnosed AML patients were obtained at the National University Hospital in Singapore with informed consent. All the AML samples were obtained through an approved protocol from the university institutional review board and follow the Helsinki protocol. Primary AML cells were cultured in IMDM with 10% FBS, FLT3 ligand (20 ng/mL), SCF (20 ng/mL), IL-3 (20 ng/mL), G-CSF (50 ng/mL), thrombopoietin (TPO; 50 ng/mL), and 1% penicillin/streptomycin. Normal CD34+CD38− hematopoietic stem/progenitor cells were purchased from Lonza AG.
Chemical reagents
DZNep was acquired from the National Cancer Institute (NCI; Rockville, MD). CM-H2DCFDA was purchased from Invitrogen (C6827). N-acetyl-l-cysteine (NAC) was obtained from Sigma-Aldrich.
DZNep treatment
Leukemia cells were seeded in 5 × 105/mL in 4-mL volume in a 6-well plate. Cells were treated with a range of concentration of DZNep (0.5, 1, 5, or 10μM). After 48 hours, cells were harvested for apoptosis assay measured by annexin V/propidium iodide (PI) flow cytometry.
Apoptosis assays
Two million cells were stained with annexin V–FITC and PI according to the manufacturer's instruction (BD Pharmingen) and analyzed using an LSR2 flow cytometer (BD Biosciences), and FlowJo software (TreeStar Inc). For the DNA fragmentation assay, DNA was extracted using the DNeasy Blood and Tissue Kit (QIAGEN). The DNA samples were separated using 2% agarose gel electrophoresis and visualized by staining with ethidium bromide.
Western blot analysis
AML cell lines or primary AML patient cells were treated with different DZNep doses for 48 hours. Cells were harvested and lysed with radioimmunoprecipitation assay (RIPA) buffer (20mM HEPES at pH 7.4, 1% Triton X-100,150mM NaCl, 1mM EDTA, 1mM EGTA, and 1× protease arrest). Total protein concentrations were determined using the Bio-Rad protein assay (Bio-Rad Laboratories Inc) and SDS-PAGE and immunoblot analyses performed with different Abs. Abs used were as follows: anti-EZH2, cleaved poly (ADP-ribose) polymerase (PARP) from Cell Signaling Technology, anti-Suz12 (07-379), anti-H3K27me3 (07-449) from Millipore, and anti-actin from Santa Cruz Biotechnology.
Affymetrix microarray analysis
MOLM-14 cells were treated with DMSO and DZNep 2μM for 24 hours. Cells were harvested and total RNA was extracted using the RNeasy Midi Kit, according to the manufacturer's instruction (QIAGEN). RNA quantity, quality, and purity were assessed with the use of the RNA 6000 Nano assay on the Agilent 2100 Bioanalyzer (Agilent Technologies).
Gene expression profiling was performed using the Affymetrix U133plus2.0 gene chip (Affymetrix) according to the manufacturer's protocol. The scanned data were processed using MicroArray Suite Version 5.0 (MAS; Affymetrix). The gene expression data were then log-transformed, median centered, and analyzed using Genespring GX 7.3.1 software (Agilent Technologies). Sequential filtering was used to select genes for subsequent analysis. First, nonexpressed probe sets (assigned an absent or marginal flag by MAS) were excluded. The remaining probe sets were subjected to ANOVA across the samples. The probe sets with significant variation with a corrected P value of < .05 after multiple testing corrections using the Benjamini and Hochberg methods were used for subsequent comparative analysis.
The gene lists were also subjected to network analysis using a web-based software MetaCore (Genego Inc). Metacore contains an interactive, manually annotated database derived from publications on proteins and small molecules that allows for representation of biologic functionality and integration of functional, molecular, or clinical information. All microarray data are available on the Gene Expression Omnibus (GEO) under accession number GSE30315.
Measurement of ROS production
CM-H2DCFDA was used as a cell-permeate indicator for intracellular ROS measurement. MOLM-14 cells were treated with DMSO control, DZNep 2μM, or antioxidant N-acetyl-l-cysteine (NAC, 20mM) for 1 hour, followed by DZNep 2μM for additional 48 hours. Cells were centrifuged and resuspended in 1× PBS containing 10μM (final concentration) CM-H2DCFDA at 37°C for 30 minutes. Cells were washed with 1× PBS, then analyzed using FITC channel in a flow cytometer.
Trx activity assay
The thioredoxin (Trx) “insulin-reducing assay” was carried out with modifications.24 Briefly, MOLM-14 cells were grown for 48 hours in complete medium containing 2μM DZNep or DMSO. Cells were harvested and lysed. Then, the protein concentration in the cell lysate samples was adjusted to 30 μg of protein in a final volume of 34 μL and preincubated with 1 μL of 2mM DTT in 50mM HEPES, pH 7.6, 1mM EDTA, 1 mg/mL BSA at 37°C in a waterbath for 15 minutes. The samples were then incubated with a 20-μL reaction mixture (200 μL of 1M HEPES pH 7.6, 40 μL of 0.2M EDTA, 40 μL of NADPH 40 mg/mL, 500 μL of insulin 10 mg/mL, and 0.5 U of Trx reductase) at 37°C for 20 minutes. The reaction was terminated by the addition of 250 μL of 6M guanidine HCl, 1mM DTNB in 0.2M Tris-HCl pH 8.0). For each sample, water was used to replace Trx reductase as negative control. Absorbance was measured at 412 nm in a Bio-Rad microplate reader. Absorbance of the negative control was subtracted from that of the corresponding sample. All the reagents were purchased from Sigma-Aldrich.
Separation of CD34+CD38− cells from primary AML patient samples
Mononuclear cells from primary AML samples obtained after informed consent were separated by Ficoll density centrifugation. CD34+CD38− subpopulations enriched for LSCs are isolated using magnetically activated cell sorting kits (Miltenyi Biotec). Briefly, leukemia mononuclear cells were washed in PBS with 2mM EDTA and 0.1% FBS, incubated with allophycocyanin-conjugated anti–human CD34 and FITC-conjugated anti–human CD38, then incubated with anti-FITC microbeads (Miltenyi Biotec). After negative immunomagnetic selection, CD38− cells were incubated with anti-allophycocyanin microbeads and positively selected through a new column. The purity of the CD34+CD38− was assessed by FACS analysis.
Methylcellulose CFU assays
CFU assays were performed using MACS HSC-CFU media (complete with Epo; Miltenyi Biotec). CD34+CD38− leukemia or normal BM cells (Lonza) were plated at 1 × 103/mL with DZNep 0.5, 1, 2μM, or DMSO control. After a 2-week incubation in a CO2 incubator, the plates were scored as positive or negative for CFU-E, CFU-G, CFU-M, CFU-GM, and CFU-GEMM.
EZH2 and TXNIP knockdown
The knockdown of EZH2 was described previously.16 Briefly, the EZH2 siRNA 5′-ACUCUGAAUGCAGUUGCU-3′ or a nontargeting control siRNA was cloned into the pSIREN-RetroQ retroviral expression vector (BD Biosciences) according to the manufacturer's instruction. Cells were infected with viruses for 72 hours. The human pLKO.1 lentiviral TXNIP shRNA target gene set was purchased from Open Biosystems. The scramble shRNA plasmid (Addgene plasmid 1864) was obtained from Addgene. MOLM-14 cells were transduced with TXNIP shRNA lentivirus pool or scramble shRNA lentivrus.
Transfection
Human full-length TXNIP cDNA was obtained from Open Biosystems and cloned into lentivirus pLVX-puro vector (Clontech) within EcoRI/BamHI site. The construct was validated by sequencing. The production and harvest of high-titer lentivirus was described previously.25 MOLM-14 cells were infected with virus particulars for 72 hours before experiments.
ChIP assays
ChIP assays were carried out using the Magna ChIP kit from Millipore as described by the manufacturer. Briefly, log-phase growing MOLM-14 cells were treated with DMSO control or DZNep 2μM for 48 hours. Cells were cross-linked with 1% formaldehyde for 10 minutes at room temperature. The chromatin was diluted with lysis buffer in the presence of a protease inhibitor cocktail, and sheared by sonication to an average of 200-1000 bp. The sheared chromatin was divided into equal amount for immunoprecipitation with either anti-Suz12 (Millipore, 07-379), anti-H3K27me3 (Millipore, 07-449) or rabbit normal IgG (negative control) with magnetic beads provided in the kit. The immunoprecipitates were eluted, reversed cross-linked, and treated with proteinase K. Purified DNA was subjected to real-time quantitative PCR (RQ-PCR) with primers specific for a region (−250 to −121 relative to transcription start site) in human the TXNIP promoter (Transcriptional Regulatory Element Database, accession number 1123). The sequences of the PCR primers used are as follows: forward primer, 5′-TCCTTCCTCTCCTTCCTCTCCTT-3′; reverse primer, 5′-TTGTTTACCAGGAGCCCGAC CAA-3′.
Real-time qRT-PCR
To quantify TXNIP gene expression, RNA samples were extracted from 5 healthy controls, 6 AML cell lines (TF-1, THP-1, MV4-11, MOLM-14, KG-1, HL60) and 5 AML patients. Typically, 1 μg of total RNA was used to generate cDNA by using SuperScript III RT (Invitrogen) with oligo-dT primer. Quantitative RT-PCR (qRT-PCR) was performed using the Power SYBR Green PCR Master Mix as recommended by the manufacturer (Applied Biosystems). GAPDH was used as the internal control. SDS 2.2.1 software (Applied Biosystems) was used to perform relative quantification of target genes using the comparative CT (ΔΔCT) method. The primer sequences were described in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Evaluation of DZNep in vivo efficacy in leukemia-engrafted mice
Subcutaneous implant model.
Exponentially growing MOLM-14 cells (5 × 106) were subcutaneously injected into loose skin between the shoulder blades and left front leg of recipient mice as described previously.26 Treatment was started when the mice had palpable tumor of 300-400 mm3 average size. DZNep was IP injected at 2 mg/kg daily. Vehicle control mice were received 1× PBS. Each group has 7 mice. The length (L) and width (W) of the tumor were measured with calipers, and tumor volume (TV) was calculated as TV = (L × W2)/2.
BM engraftment model.
Six- to 8-week-old female NOD/SCID mice were purchased from Animal Resource Center and maintained in specific pathogen-free conditions. As a standard procedure to improve the engraftment efficiency, mice were given Endoxan (cyclophosphamide; Baxter Oncology GmbH) 150 mg/kg/d for 2 days followed by 1 rest day before leukemia cells were injected.26 MOLM-14 cells (2 × 106) were injected into the tail vein of the mice. Mice were treated with either sterile PBS as control or DZNep 2 mg/kg/d by IP in 200-μL volume from the second day. Seven mice were in each group.
The protocol was reviewed and approved by the Institutional Animal Care and Use Committee at the National University of Singapore in compliance with the guidelines on the care and use of animals for scientific purpose.
Statistical analysis
Differences among values obtained from tumor volume reduction or in vitro experiments of the treatment groups were compared with the untreated control group by Student t test, and P values of < .05 were considered to be significant. Survival analysis was performed by Kaplan-Meier analysis (Version 12; SPSS). Survival curves of the different treatment groups were compared using the log-rank test (P < .05).
Results
DZNep showed potent anti-leukemia effect in vitro and in vivo
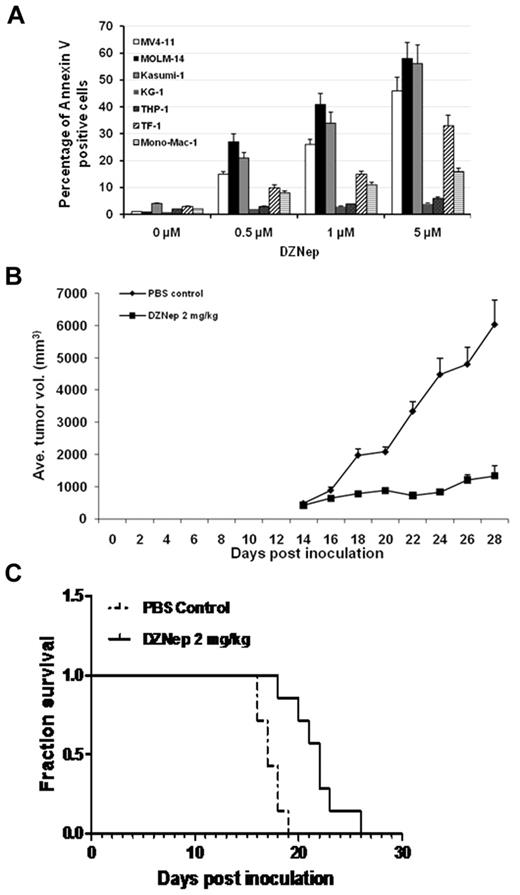
Initial studies were performed to screen the effect of DZNep on a panel of AML cell lines. Cells were treated with DMSO or DZNep at concentrations of 500nM, 1μM, and 5μM for 48 hours in suspension culture, followed by apoptosis assay by flow cytometry. Three groups of cell lines with different sensitivity were discernible. With 5μM DZNep, MV4-11, MOLM-14, and Kasumi-1 were very sensitive to DZNep with > 40% of apoptotic cells. Mono-Mac-1 and TF-1 showed moderate response to DZNep with < 40% but > 15% of apoptotic cells. THP-1 and KG-1 were relatively resistant to DZNep with < 10% apoptotic cells (Figure 1A, Table 1).
DZNep showed potent anti-AML efficacy in vitro and in vivo. (A) AML cell lines were treated with either DMSO control or DZNep at dose of 0.5, 1, and 5μM for 48 hours. Cells were harvested, washed, and stained with annexin V/PI double dye, then subjected to flow cytometric analysis. (B) Mice were treated with PBS control or DZNep 2 mg/kg/d, respectively. Tumor volume curves were constructed with measurements taken by caliper. Average of tumor volume was calculated as the average of 7 mice in each group ± SD. (C) Survival analysis showed that DZNep treatment improved the survival time of mice inoculated with human leukemia (P < .05). Seven mice in each group were used for the construction of the survival curves.
DZNep showed potent anti-AML efficacy in vitro and in vivo. (A) AML cell lines were treated with either DMSO control or DZNep at dose of 0.5, 1, and 5μM for 48 hours. Cells were harvested, washed, and stained with annexin V/PI double dye, then subjected to flow cytometric analysis. (B) Mice were treated with PBS control or DZNep 2 mg/kg/d, respectively. Tumor volume curves were constructed with measurements taken by caliper. Average of tumor volume was calculated as the average of 7 mice in each group ± SD. (C) Survival analysis showed that DZNep treatment improved the survival time of mice inoculated with human leukemia (P < .05). Seven mice in each group were used for the construction of the survival curves.
The IC50 of DZNep in cell lines and 16 AML patients with clinical characteristics
| U. Pt no. and cell lines . | Age, y/sex . | FAB subtype . | Karyotype . | MLL rearrangement . | FLT3-ITD . | FLT3-TKD . | DZNep IC50, μM . |
|---|---|---|---|---|---|---|---|
| 1 | 34/F | M5a | N | Neg | ND | ND | 8.1 |
| 2 | 68/M | M1/M5 | +11 | Neg | Neg | Neg | 12.0 |
| 3 | 28/M | M1 | +5p15 | Pos | Neg | Neg | 9.5 |
| 4 | 68/F | M1 | −13q21 | Pos | Neg | Neg | 11.5 |
| 5 | 36/M | M2 | N | Neg | Neg | Pos | 7.0 |
| 6 | 37/F | M5 | N | Neg | Neg | Pos | 6.0 |
| 7 | 39/M | M1 | t(6;9)(p23;q34) | Neg | Pos | Neg | 5.2 |
| 8 | 69/F | M1 | t(9;22)(q34;q11), +8, +19 | Neg | Neg | Neg | 15.0 |
| 9 | 33/F | M4 | inv(16)(p13q22) | Neg | ND | ND | 12.9 |
| 10 | 68/F | M2 | N | Neg | ND | Neg | 9.3 |
| 11 | 56/M | ND | t(9;22)(q34;q11), +8p22, −15, −18 | Neg | Neg | Neg | 14.8 |
| 12 | 58/F | M4 | N | Neg | ND | ND | 10.0 |
| 13 | 43/M | M2 | t(8;21)(q22;q22) | Neg | Neg | Neg | 3.7 |
| 14 | 49/M | ND | ND | ND | Pos | Neg | 3.5 |
| 15 | 42/F | M1 | N | Neg | Pos | Neg | 4.6 |
| 16 | 59/F | M2 | N | Neg | ND | ND | 5.8 |
| MOLM-14 | NA | M5 | +6, +8, ins(11;9)(q23;p22p23 | Pos | Pos | Neg | 4.2 |
| MV4–11 | NA | M5 | +8, +19, t(4;11)(q21;q23) | Pos | Pos | Neg | 6.3 |
| Kasumi-1 | NA | M2 | t(8:21)(q22;q22) | Neg | Neg | Neg | 4.8 |
| TF-1 | NA | M6 | Hyperdiploid | Neg | Neg | Neg | 12.5 |
| Mono-Mac-1 | NA | M5 | t(9;11)(p22;q23) | Pos | Neg | Neg | 15.0 |
| THP-1 | NA | M5 | t(9;11)(p22;q23) | Pos | Neg | Neg | Resistant |
| KG-1 | NA | M1 | Hypodiploid | Neg | Neg | Neg | Resistant |
| U. Pt no. and cell lines . | Age, y/sex . | FAB subtype . | Karyotype . | MLL rearrangement . | FLT3-ITD . | FLT3-TKD . | DZNep IC50, μM . |
|---|---|---|---|---|---|---|---|
| 1 | 34/F | M5a | N | Neg | ND | ND | 8.1 |
| 2 | 68/M | M1/M5 | +11 | Neg | Neg | Neg | 12.0 |
| 3 | 28/M | M1 | +5p15 | Pos | Neg | Neg | 9.5 |
| 4 | 68/F | M1 | −13q21 | Pos | Neg | Neg | 11.5 |
| 5 | 36/M | M2 | N | Neg | Neg | Pos | 7.0 |
| 6 | 37/F | M5 | N | Neg | Neg | Pos | 6.0 |
| 7 | 39/M | M1 | t(6;9)(p23;q34) | Neg | Pos | Neg | 5.2 |
| 8 | 69/F | M1 | t(9;22)(q34;q11), +8, +19 | Neg | Neg | Neg | 15.0 |
| 9 | 33/F | M4 | inv(16)(p13q22) | Neg | ND | ND | 12.9 |
| 10 | 68/F | M2 | N | Neg | ND | Neg | 9.3 |
| 11 | 56/M | ND | t(9;22)(q34;q11), +8p22, −15, −18 | Neg | Neg | Neg | 14.8 |
| 12 | 58/F | M4 | N | Neg | ND | ND | 10.0 |
| 13 | 43/M | M2 | t(8;21)(q22;q22) | Neg | Neg | Neg | 3.7 |
| 14 | 49/M | ND | ND | ND | Pos | Neg | 3.5 |
| 15 | 42/F | M1 | N | Neg | Pos | Neg | 4.6 |
| 16 | 59/F | M2 | N | Neg | ND | ND | 5.8 |
| MOLM-14 | NA | M5 | +6, +8, ins(11;9)(q23;p22p23 | Pos | Pos | Neg | 4.2 |
| MV4–11 | NA | M5 | +8, +19, t(4;11)(q21;q23) | Pos | Pos | Neg | 6.3 |
| Kasumi-1 | NA | M2 | t(8:21)(q22;q22) | Neg | Neg | Neg | 4.8 |
| TF-1 | NA | M6 | Hyperdiploid | Neg | Neg | Neg | 12.5 |
| Mono-Mac-1 | NA | M5 | t(9;11)(p22;q23) | Pos | Neg | Neg | 15.0 |
| THP-1 | NA | M5 | t(9;11)(p22;q23) | Pos | Neg | Neg | Resistant |
| KG-1 | NA | M1 | Hypodiploid | Neg | Neg | Neg | Resistant |
AML indicates acute myeloid leukemia; U. Pt no., unique patient number; FAB, French-American-British; N, normal karyotype; MLL, mixed lineage leukemia gene; FLT3, FMS-like tyrosine kinase 3; ITD, internal tandem duplication; TKD, tyrosine kinase domain mutation; NA, not applicable; ND, not done; Neg, negative; and Pos, positive.
To validate the clinical relevance of the results observed in cell lines, primary cells from 16 patients with AML were incubated with DMSO, DZNep 2, 5, or 10μM for 48 hours, followed by FACS analysis using annexin V/PI double staining to detect apoptotic cells. Robust, dose-dependent apoptosis by DZNep treatment was observed in primary AML samples. The IC50 of DZNep and clinical characteristics of patients were summarized in Table 1. To investigate whether DZNep treatment would exert an in vivo anti-leukemia effect, we conducted subcutaneous implant and BM engraftment animal experiments.
DZNep-depleted PRC2 and inhibited H3K27me3 in AML cell lines and primary AML cells
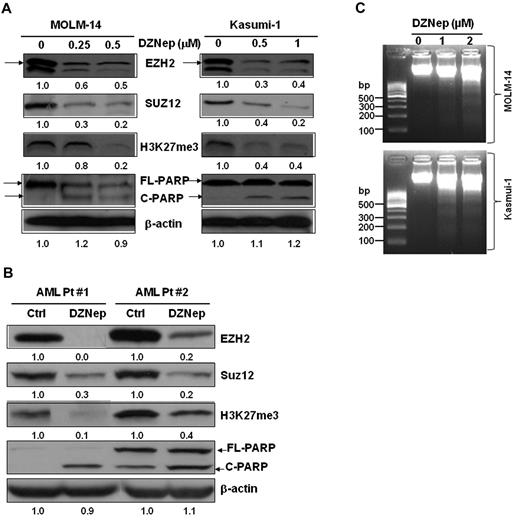
We and others previously reported that DZNep effectively depleted PRC2 proteins EZH2, Suz12, and EED and inhibited H3K27me3 in breast cancer cells.16,20,21 Consistent with the action in breast cancer, treatment with DZNep potently decreased the protein expression of EZH2, SUZ12, and inhibited H3K27me3 in MOLM-14 and Kasumi-1 cells (Figure 2A), as well as in primary AML cells from 2 patients (Figure 2B). In agreement with the apoptosis assays by FACS analysis (Figure 1), cleaved PARP, a hallmark of apoptosis, was significantly induced after treatment with DZNep (Figure 2A-B), indicating leukemia cells undergoing apoptosis. We further showed that DZNep treatment induced internucleosomal DNA fragmentation, a key feature of apoptosis, in both MOLM-14 and Kasumi-1 cells (Figure 2C).
DZNep decreased the protein expression of EZH2, Suz12, H3K27me3, and induced cleaved PARP in leukemia cells. (A) MOLM-14 and Kasumi-1 cells were incubated with 0.1% DMSO, DZNep 0.25μM, 0.5μM, and 0.1% DMSO, 0.5μM, 1μM, respectively, for 48 hours. (B) Primary AML cells were treated with DZNep 5μM for 48 hours. Cells were harvested, lysed, and subjected to immunoblot analysis with primary Abs indicated. β-actin was used as loading controls. Densitometric analysis was performed using Amersham Image Scanner with LabScan ImageQuant TL Software (Amersham Biosciences). The expression ratio of EZH2, Suz12, and H3K27me3 was calculated as the DZNep-treated samples relative to control samples after normalization with respective β-actin level. Arrows indicate EZH2; C, cleaved; and FL (full-length), PARP protein, respectively. (C) Effect of DZNep on DNA fragmentation in MOLM-14 and Kasumi-1 cells.
DZNep decreased the protein expression of EZH2, Suz12, H3K27me3, and induced cleaved PARP in leukemia cells. (A) MOLM-14 and Kasumi-1 cells were incubated with 0.1% DMSO, DZNep 0.25μM, 0.5μM, and 0.1% DMSO, 0.5μM, 1μM, respectively, for 48 hours. (B) Primary AML cells were treated with DZNep 5μM for 48 hours. Cells were harvested, lysed, and subjected to immunoblot analysis with primary Abs indicated. β-actin was used as loading controls. Densitometric analysis was performed using Amersham Image Scanner with LabScan ImageQuant TL Software (Amersham Biosciences). The expression ratio of EZH2, Suz12, and H3K27me3 was calculated as the DZNep-treated samples relative to control samples after normalization with respective β-actin level. Arrows indicate EZH2; C, cleaved; and FL (full-length), PARP protein, respectively. (C) Effect of DZNep on DNA fragmentation in MOLM-14 and Kasumi-1 cells.
DZNep differentially impacts colony formation of leukemia and normal HSCs
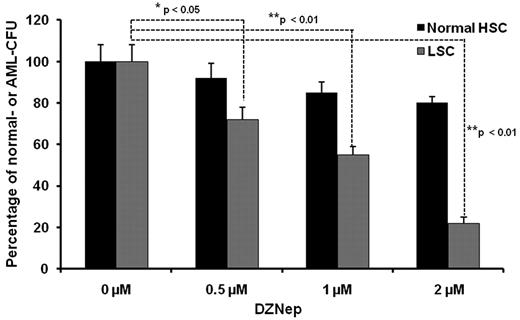
It has been demonstrated that AML is a hierarchical LSC disease, originating from a primitive hematopoietic cell. LSCs are enriched in the CD34+CD38− fraction.27 As inhibition of EZH2 and H3K27me3 may affect stem cell maintenance and differentiation in both normal stem cells and LSCs, we wanted to examine whether DZNep exerted a differential effect between normal HSCs and LSCs. A total 1 × 103 of CD34+CD38− cells were cultured in MACS HSC-CFU media in the presence of cytokines with 0.1% DMSO (control), 0.5μM, 1μM, and 2μM DZNep for 14 days. For the normal BM samples, total colony numbers were calculated as the sum of BFU-Es, CFU-Es, CFU-GMs, CFU-GEMMs, CFU-Ms, and CFU-Gs. For the AML BM samples, the colonies were usually less differentiated and distinguished, so we counted the total number of colonies as AML-CFU if the colonies consisted of > 10 cells. Treatment with DZNep at 0.5, 1, and 2μM decreased the number of AML-CFUs by 28%, 45%, and 78%, respectively, but minimally inhibited the colony formation of normal samples (Figure 3). This differential effect suggests that DZNep may specifically target LSC-enriched subpopulations, while potentially sparing normal HSCs.
The effect of DZNep on colony formation of LSCs from AML patients and normal HSCs. Methycellulose-based colony assay performed in CD34+CD38− BM cells form 3 AML patients and 3 healthy donors. Each data point represents the average of 3 samples with triplicate platings normalized to DMSO control. The data were analyzed by the Student t test comparing normal HSCs vs LSCs (*P < .05, **P < .01). All the AML samples were obtained through an approved protocol from the university institutional review board and follow the Helsinki protocol.
The effect of DZNep on colony formation of LSCs from AML patients and normal HSCs. Methycellulose-based colony assay performed in CD34+CD38− BM cells form 3 AML patients and 3 healthy donors. Each data point represents the average of 3 samples with triplicate platings normalized to DMSO control. The data were analyzed by the Student t test comparing normal HSCs vs LSCs (*P < .05, **P < .01). All the AML samples were obtained through an approved protocol from the university institutional review board and follow the Helsinki protocol.
Transcriptome analysis reveals up-regulation of TXNIP and down-regulation of endoplasmic reticulum (ER) stress-related survival genes
Different from the previous studies16,21 that delineated transcriptional changes at 72 hours after DZNep treatment in breast cancer, we focused on the early gene alterations induced by DZNep at 24 hours in AML. We found 239 genes differentially expressed by > 2-fold (91 up and 148 down, supplemental Table 2) between DMSO control and DZNep-treated MOLM-14 cells using the Affymetrix array platform. The top 20 elevated and repressed genes were depicted in supplemental Figure 1A, arranged according to average fold changes. We found that TXNIP (also known as thioredoxin-binding protein 2 [TBP-2] or vitamin D3 up-regulated protein 1 [VDUP1]),28 was among the top 3 up-regulated genes (5-fold) on DZNep treatment (supplemental Figure 1A, indicated by arrow).
To gain an overview of genes modulated by DZNep treatment, we searched for enrichment of functional gene ontology (GO) annotations among genes that are at least 2-fold differentially expressed (supplemental Figure 1B). In total, 30 biologic functions were significantly associated with response of treatment. Overrepresented GO terms included those related to the regulation of cell proliferation and development process, regulation of apoptosis and cell cycle, immune response, metabolic process, as well as response to stress. The GO terms analysis results were consistent with known changes in gene expression induced by epigenetic modulation of DZNep. But it also revealed some potentially novel function of DZNep such as mediating stress response.
Next, we used the MetaCore program to map the direct interactions among the differentially expressed genes. Notably, several genes clustered around DDIT3 (DNA-damage-inducible transcript 3, also known as C/EBP zeta, or GADD1530), which was associated with DNA damage pathway (supplemental Figure 1C). A cluster of ER stress-regulated genes centered on HSPA5 (heat shock 70 kDa protein 5, also known as GRP78 [glucose-regulated protein, 78 kDa]; BiP, Ig heavy chain-binding protein) and PERK (PRKR-like endoplasmic reticulum kinase, also known as eIF2AK3 [eukaryotic translation initiation factor 2 α kinase 3]) also appeared on the network map (supplemental Figure 1D). Finally, we manually searched the gene list and classified 24 (16%) ER stress-related genes among the list of down-regulated genes (supplemental Figure 1E). These ER stress-related genes, such as GRP78, GRP94, PERK, PDIA isoform 3, 4, and 5, are usually associated with cell survival; however, all were reduced by DZNep treatment.
DZNep increased the expression of TXNIP at the mRNA and protein levels, which resulted in decreasing Trx activity, elevating ROS production, and targeting ER stress
TXNIP, first cloned from HL60 stimulated with vitamin D3, antagonizes Trx, a redox-active protein, and is part of a major system governing the balance of cellular ROS.28,29 It has been demonstrated that regulation of ROS is essential for HSC self-renewal and maintenance of the HSC pool,30-32 so we decided to further study TXNIP.
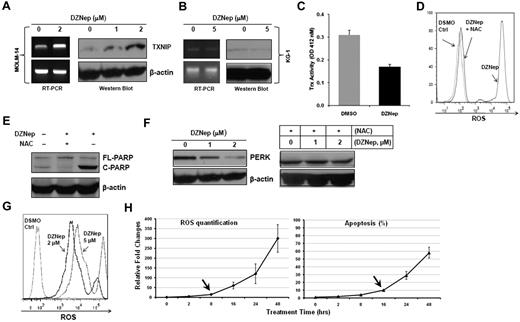
We first validated the induced expression of TXNIP mRNA by qRT-PCR (6-fold increase) and conventional RT-PCR (Figure 4A). Next, we demonstrated that DZNep dose-dependently elevated the expression of TXNIP at the protein level (Figure 4A) in MOLM-14 cells. In contrast, DZNep treatment failed to augment the expression of TXNIP in KG-1 cells, which were resistant to DZNep (Figure 4B). This validated our gene expression results and suggested the induction of TXNIP by DZNep is specific to sensitive cell lines and may be important in mediating sensitivity to DZNep. Considering that TXNIP plays an important role in the regulation of both redox state and oxidative stress,29 we set out to determine the changes of Trx activity and ROS production caused by DZNep treatment. In MOLM-14 cells incubated with DZNep, Trx activity was decreased ∼ 2-fold (Figure 4C). MOLM-14 cells treated with 2μM DZNep for 48 hours showed a remarkable increase in the accumulation of ROS compared with cells cultured with vehicle control (Figure 4D). Pretreatment with 20mM NAC, a ROS scavenger, 1 hour before adding DZNep led to almost complete inhibition of intracellular ROS generation (Figure 4D). Importantly, NAC also blocked DZNep-induced apoptosis in MOLM-14, as evidenced by reduced cleaved PARP (Figure 4E), suggesting that induction of apoptosis after DZNep treatment is the result of reduction of Trx activity and increased ROS production. We further validated the induction of TXNIP protein and ROS generation by DZNep treatment in 2 additional AML cell lines (MV4-11 and Kasumi-1) and 2 AML patient samples (supplemental Figure 2).
DZNep treatment increased TXNIP expression and ROS production. TXNIP expression was determined by RT-PCR and Western blot analysis in MOLM-14 (A) and KG-1 (B) cells treated with either DMSO control or DZNep for 24 hours. (C) Trx activity in MOLM-14 cells treated with DMSO control or 2μM DZNep for 48 hours was measured at 412nM. Three separate measurements were indicated as mean ± SD. (D) FACS analysis of ROS production in MOLM-14 cells treated with DMSO control, DZNep, or NAC + DZNep. (E) Western blot analysis of cleaved PARP. (F) MOLM-14 cells were treated with DMSO control, DZNep 1μM or 2μM for 48 hours with or without NAC. Cells were washed, lysed, and subjected to Western blot analysis of PERK. β-actin was used as a loading control. (G) One representative FACS profile of ROS production in one CD34+CD38− leukemia early progenitor cells treated with DZNep. (H) FACS analysis of ROS production (left panel) and apoptosis (right panel) in MOLM-14 cells treated with DZNep 2μM in a time-dependent manner. The amount of ROS production and apoptosis in treated samples were normalized to that of baseline in the control samples (0 hours). Three separate measurements were indicated as mean ± SD. Arrows indicate the starting time point when ROS and apoptosis were significantly increased, respectively.
DZNep treatment increased TXNIP expression and ROS production. TXNIP expression was determined by RT-PCR and Western blot analysis in MOLM-14 (A) and KG-1 (B) cells treated with either DMSO control or DZNep for 24 hours. (C) Trx activity in MOLM-14 cells treated with DMSO control or 2μM DZNep for 48 hours was measured at 412nM. Three separate measurements were indicated as mean ± SD. (D) FACS analysis of ROS production in MOLM-14 cells treated with DMSO control, DZNep, or NAC + DZNep. (E) Western blot analysis of cleaved PARP. (F) MOLM-14 cells were treated with DMSO control, DZNep 1μM or 2μM for 48 hours with or without NAC. Cells were washed, lysed, and subjected to Western blot analysis of PERK. β-actin was used as a loading control. (G) One representative FACS profile of ROS production in one CD34+CD38− leukemia early progenitor cells treated with DZNep. (H) FACS analysis of ROS production (left panel) and apoptosis (right panel) in MOLM-14 cells treated with DZNep 2μM in a time-dependent manner. The amount of ROS production and apoptosis in treated samples were normalized to that of baseline in the control samples (0 hours). Three separate measurements were indicated as mean ± SD. Arrows indicate the starting time point when ROS and apoptosis were significantly increased, respectively.
Accumulating evidence suggests that a vicious cycle exists between ER stress and ROS production.33 Severe ER stress ultimately leads to activation of cell-death pathway.34 Consistent with this notion, the high amount of ROS induced by DZNep treatment targeted the ER stress-response pathway, repressing a cluster of prosurvival genes (supplemental Figure 1F), and promoting apoptosis. In agreement with the gene expression data, Western blot analysis showed that DZNep treatment dose-dependently reduce PERK protein, a serine/threonine protein kinase. However, this reduction was abrogated by NAC, suggesting the induction of ER stress and down-regulation of prosurvival genes may be downstream of ROS production (Figure 4F). To test this mechanism is not limited to MOLM-14 cells, we examined the expression of 7 genes including TXNIP, PERK, HYOU1, DDIT3, HSPA5, PDAI3, GRP94 in MV4-11, Kasumi-1 and 2 AML patient BM cells after DZNep treatment. qRT-PCR analysis showed that consistent with the result in MOLM-14, DZNep treatment up-regulated the expression of TXNIP, while decreasing ER stress-related genes in 2 AML cell lines and 2 AML BM cells (supplemental Table 3; see “Methods” in supplemental Table 2 for primer sequences).
We next asked whether DZNep treatment also induced ROS production in AML LSC-enriched subpopulations. To that end, 3 CD34+CD38− samples isolated from primary AML BM samples were incubated with DMSO, DZNep 2μM and 5μM for 48 hours. DZNep dose-dependently increased ROS production 300- to 500-fold in these 3 samples. One representative ROS profile was shown in Figure 4G. To determine the sequential occurrence of EZH2 depletion, ROS production, and apoptosis, we treated MOLM-14 cells with DZNep 2μM for 0, 8, 16, 24, and 48 hours. As shown in supplemental Figure 3A, although DZNep inhibited EZH2 as early as 2 hours, down-regulation of H3K27m3 was observed at 8 hours, and increased TNXIP was seen at even later time point (16 hours). Consistent with this, FACS analysis showed that DZNep induced the first significant increase in ROS production from 8 to 16 hours before the first significant increase in cell apoptosis from 16 to 24 hours (Figure 4H, supplemental Figure 3B for primary FACS plots). These sequential changes indicate the induction of apoptosis is the consequence of EZH2 depletion, TXNIP induction, and ROS production by DZNep treatment.
Silencing of EZH2 increased TXNIP and ectopic expression of TXNIP induced ROS production and cell death
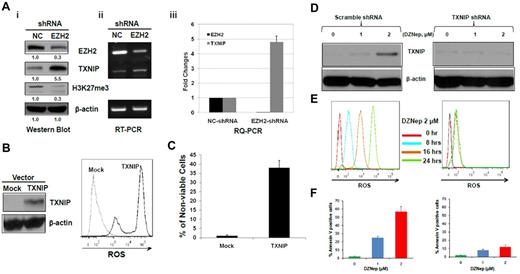
To determine whether the phenotypic and molecular consequences observed after DZNep treatment is predominantly mediated through EZH2 silencing and not other targets, we performed experiments to specifically knockdown EZH2 in MOLM-14 cells. Western blot (Figure 5Ai) and RT-PCR (Figure 5Aii) confirmed the specific inhibition of EZH2, and qRT-PCR (Figure 5Aiii) revealed ∼ 33-fold reduction of EZH2 expression by pSIREN-EZH2 shRNA compared with nontargeting shRNA control. Knockdown of EZH2 led to increased expression of TXNIP at both mRNA (Figure 5Ai) and protein level (Figure 5Aii-iii), with concomitant depletion of H3K27me3 (Figure 5Ai). To further verify that the induction of ROS is related to TXNIP overexpression and not nonspecific effect of DZNep on the mitochondria, we overexpressed TXNIP in MOLM-14 cells and determined whether similar induction of ROS and apoptosis as seen with DZNep was observed. Cells transfected with pLVX-TXNIP significantly increased ROS production and cell death compared with MOLM-14 cells transfected with mock vector (Figure 5B-C). To further demonstrate the critical role of TXNIP, we performed shRNA knockdown experiments and then treated cells with DZNep, followed by examining ROS production and apoptosis in these 2 cell lines. Knockdown of TXNIP effectively blocked ROS production by DZNep after 8 hours and largely abrogated the increased apoptosis induced by DZNep after 48 hours (right panels of Figure 5D-F), but the scramble shRNA failed to do so (left panels of Figure 5D-F).
The effect of knockdown of EZH2 or ectopic expression of TXNIP or knockdown of TXNIP in MOLM-14 cells. (A) MOLM-14 cells were transduced with pSIREN-EZH2 shRNA or nontargeting virus for 72 hours. The protein levels of EZH2, TXNIP, and H3K27me3 were determined by Western blot analysis. (i) β-actin was used as the loading control; the mRNA levels of EZH2 and TXNIP were analyzed by (ii) RT-PCR and (iii) RQ-PCR. Densitometric analysis was performed using Amersham Image Scanner with LabScan ImageQuant TL software. The expression ratio of EZH2, and TXNIP was calculated as the EZH2-shRNA–treated samples relative to control samples after normalization with respective β-actin level. (B) MOLM-14 cells were transfected with pLVX-TXNIP or control lentivirus (Mock) for 72 hours, followed by Western blot analysis of TXNIP and FACS analysis of ROS production. (C) Trypan blue exclusion method was used to determine the cell viability in a hemocytometer under an invert microscope. (D-F) MOLM-14 cells were transduced with scramble- (left panels) or TXNIP-shRNA (right panels). Cells were then incubated with either DZNep 0, 1, 2μM, followed by Western blot analysis of TXNIP protein (D), and apoptosis after 48 hours (F), or DZNep 2μM, followed by quantification of ROS production at 0, 8, 16, and 24 hours (E). Three separate determinations were indicated as mean ± SD.
The effect of knockdown of EZH2 or ectopic expression of TXNIP or knockdown of TXNIP in MOLM-14 cells. (A) MOLM-14 cells were transduced with pSIREN-EZH2 shRNA or nontargeting virus for 72 hours. The protein levels of EZH2, TXNIP, and H3K27me3 were determined by Western blot analysis. (i) β-actin was used as the loading control; the mRNA levels of EZH2 and TXNIP were analyzed by (ii) RT-PCR and (iii) RQ-PCR. Densitometric analysis was performed using Amersham Image Scanner with LabScan ImageQuant TL software. The expression ratio of EZH2, and TXNIP was calculated as the EZH2-shRNA–treated samples relative to control samples after normalization with respective β-actin level. (B) MOLM-14 cells were transfected with pLVX-TXNIP or control lentivirus (Mock) for 72 hours, followed by Western blot analysis of TXNIP and FACS analysis of ROS production. (C) Trypan blue exclusion method was used to determine the cell viability in a hemocytometer under an invert microscope. (D-F) MOLM-14 cells were transduced with scramble- (left panels) or TXNIP-shRNA (right panels). Cells were then incubated with either DZNep 0, 1, 2μM, followed by Western blot analysis of TXNIP protein (D), and apoptosis after 48 hours (F), or DZNep 2μM, followed by quantification of ROS production at 0, 8, 16, and 24 hours (E). Three separate determinations were indicated as mean ± SD.
These results validate that the induction of apoptosis by DZNep is mainly mediated through EZH2 silencing with consequential induction of TXNIP and ROS resulting in apoptosis of AML cells.
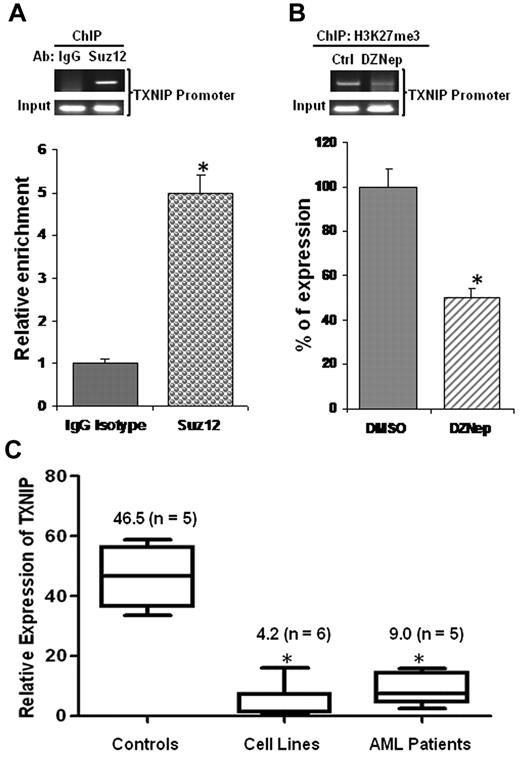
TXNIP was repressed in AML cells, its promoter enriched for PRC2 complex occupancy, and H3K27me3 reduced on DZNep treatment
To determine whether TXNIP is a direct target of PRC2, we conducted ChIP experiments using anti-Suz12 Ab to assess the PRC2 complex binding to the TXNIP promoter. We observed that the expression of TXNIP was ∼ 5-fold higher in DNA samples immunoprecipitated with anti-Suz12 Ab than that of the isotype control IgG Ab (Figure 6A), demonstrating PRC2 enrichment in the TXNIP promoter region. To test whether up-regulation of TXNIP was because of DZNep-mediated reduction of H3K27me3 at TXNIP promoter region, we performed ChIP assays using anti-H3K27me3 Ab. Quantitative RT-PCR revealed that H3K27me3 was decreased ∼ 50% in the TXNIP promoter region of DZNep-treated MOLM-14 samples compared with DMSO control samples (Figure 6B).
PRC2 complex binds TXNIP promoter and TXNIP is repressed in AML. (A) ChIP assays were performed in MOLM-14 cells using Abs against Suz12 or a negative control IgG and analyzed by RQ-PCR amplifying a region of −250 to −121 relative to transcription start site of TXNIP promoter. The relative enrichments of PRC2 complex in the TXNIP promoter region were normalized to isotype controls. The experiments were conducted in triplicates (mean ± SD). (B) MOLM-14 cells treated with DMSO control or DZNep 2μM for 48 hours. ChIP assays were conducted using Ab against H3K27me3. The relative reductions were shown in mean ± SD of triplicate measurement. (C) Real-time qRT-PCR analysis of TXNIP expression in healthy controls, AML cell lines, and primary AML patient cells. The expression of TXNIP in TF-1 cells was set as 1 (baseline), which is the lowest among the 6 cell lines tested. The average of CT value of TXNIP in TF-1 sample was 24 ± 0.5. The numbers in the brackets indicate the sample size.
PRC2 complex binds TXNIP promoter and TXNIP is repressed in AML. (A) ChIP assays were performed in MOLM-14 cells using Abs against Suz12 or a negative control IgG and analyzed by RQ-PCR amplifying a region of −250 to −121 relative to transcription start site of TXNIP promoter. The relative enrichments of PRC2 complex in the TXNIP promoter region were normalized to isotype controls. The experiments were conducted in triplicates (mean ± SD). (B) MOLM-14 cells treated with DMSO control or DZNep 2μM for 48 hours. ChIP assays were conducted using Ab against H3K27me3. The relative reductions were shown in mean ± SD of triplicate measurement. (C) Real-time qRT-PCR analysis of TXNIP expression in healthy controls, AML cell lines, and primary AML patient cells. The expression of TXNIP in TF-1 cells was set as 1 (baseline), which is the lowest among the 6 cell lines tested. The average of CT value of TXNIP in TF-1 sample was 24 ± 0.5. The numbers in the brackets indicate the sample size.
To determine whether TXNIP is indeed repressed in AML, we performed qRT-PCR to measure its mRNA in healthy controls, AML cell lines, and primary AML cells. Our result demonstrated that TXNIP expression was ∼ 11-fold and 5-fold lower in AML cell lines and primary AML cells, respectively, than its expression in healthy controls (Figure 6C).
Discussion
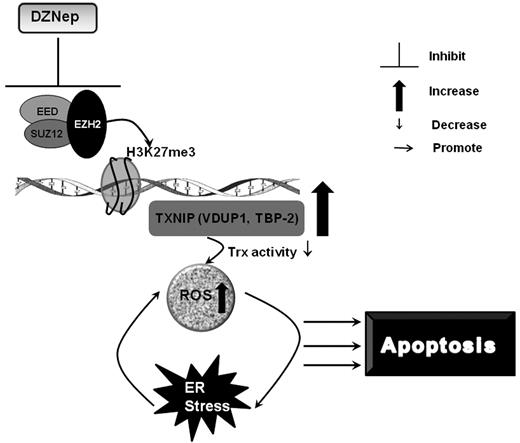
In this study, we characterized the anti-leukemia effect of the histone methyltransferase inhibitor, DZNep, against AML in vitro and in vivo. Consistent with previous studies but in a larger panel of AML cell lines, we showed that DZNep has activity in AML.22 In addition, we showed that DZNep has a differential effect on LSC-enriched subpopulations compared with normal HSCs. We further demonstrated that much of the effect of DZNep is mediated through the reactivation of TXNIP via the depletion of PRC2 proteins and associated histone (H3K27) methylation. Importantly, using gene expression profiling, shRNA, and ChIP assays, we uncovered a novel mechanism by which up-regulation of TXNIP, reduced Trx activity, and increased ROS production, resulting in apoptotic cell death (Figure 7).
A schematic summary of the mechanisms of DZNep-induced apoptosis in AML.
A schematic summary of the mechanisms of DZNep-induced apoptosis in AML.
TXNIP has been shown to be a tumor suppressor gene in human breast,35 colon, and lung cancer36 through inhibition of cell proliferation and arrest of the cell cycle at G0/G1 phase. Furthermore, TXNIP also acts as a suppressor of melanoma metastasis,36 and induces β-cell apoptosis by increasing Bax/Bcl-2 ratio.37 The strongest evidence comes from TXNIP-deficient mice associated with a higher incidence of hepatocellular carcinoma.38 It has been demonstrated in many studies that vorinostat (SAHA), the first histone deacetylase inhibitor approved by Food and Drug Administration for clinical use, exhibits potent antitumor function at least in part through reactivation of TXNIP.39 Here, we showed that TXNIP is repressed in human AML and is a direct target of PRC2-H3K27me3. Therefore, TXNIP seems to be silenced by 2 epigenetic regulators, that is, histone deacetylation and H3K27me3 in cancers. Our observations indicate that repression of TXNIP is an important step in tumor cell transformation, and therefore possibly requires both gene silencing mechanisms to ensure its tight inactivation.
TXNIP directly binds Trx and inhibits its mRNA expression and function as a major redox control molecule, thereby leading to ROS accumulation.28,29 ROS can be an initiator of ER stress or products of ER stress. Moderate increase in ROS production induces ER response, preserves cell function, and promotes cell survival in an adaptive mechanism by which cell produces more chaperone proteins, such as GRP78, GRP94, PDI, and activates PERK. Several studies demonstrated that moderately increased ER stress contributes to leukemic transformation.40,41 In particular, GRP78 has been identified as a key survival factor for cancer cell and a novel biomarker for tumor progression and clinical prognosis.42 PERK plays a major role in cell survival by abrogation of phosphorylation of eIF2alpha during ER stress.41 However, excessive amount of ROS causes severe ER stress and initiates apoptotic cascade, leading to cell death.34 As revealed by our microarray analysis, notably, DZNep treatment significantly inhibited these ER stress-response genes, withdrawing adaptive protection, which may further contributed to the anti-leukemia effect. Our analysis therefore revealed intriguing indirect evidence of a novel function of PRC2 in connection with ER stress regulation. This will need to be further validated and characterized.
Sustainable self-renew is one hallmark of HSCs and a main characteristic of LSCs.43 Primitive HSCs contain low levels of ROS and reside in the low-oxygenic BM niche.31 Several studies demonstrated that dysregulation of intracellular ROS damages self-renewal and leads to stem cell failure.30,32 Therefore, our observation of higher intracellular ROS production in the CD34+CD38− LSC-enriched subpopulations after exposure to DZNep suggests a possible mechanism for the differential effect of DZNep on LSCs compared with normal HSCs. In agreement with our findings, it was recently reported that DZNep strongly decreases glioblastoma cancer stem cell self-renewal and abrogates its tumor-initiating capacity.18
Our findings demonstrate that reactivation of TXNIP induced by either treatment with DZNep, EZH2-shRNA, or forced expression of TXNIP itself could increase ROS accumulation suggesting that DZNep-mediated depletion of EZH2 and H3K27me3 is responsible for the up-regulation of TXNIP and the ensuing biologic cascades. Taken together, our work uncovers a novel mechanism of DZNep-induced anti-leukemia efficacy and provides strong rationale to further develop strategies to target EZH2 in AML, either through DZNep or its new analogs, or other molecules currently in development.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors gratefully acknowledge Dr Gaofeng Huang for generating the heatmaps. They also thank Prof H. Philip Koeffler for critical suggestions.
This work was supported by the Singapore National Research Foundation and the Ministry of Education under the Research Center of Excellence Program. W.-J.C. is also supported by the National Medical Research Council (NMRC) Clinician Scientist Investigator award.
Authorship
Contribution: J.Z., Q.Y., and W.-J.C. conceptualized the original idea, designed the experiments, and analyzed the data; J.Z. performed the experiments and wrote the paper; J.Z., C.B., L.-L.C., S.M., S.-C.L., K.-G.T., and T.-L.K. contributed to the in vitro experiments; and J.Z. and S.-C.L. carried out the animal experiments.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Wee-Joo Chng, Associate Professor, Department of Hematology-Oncology, National University Cancer Institute of Singapore, The National University Health System (NUHS), 1E, Kent Ridge Rd, Singapore 119228; e-mail: mdccwj@nus.edu.sg.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal