Abstract
Not all individuals exposed to HIV become infected. Understanding why these HIV-exposed seronegative individuals remain uninfected will help inform the development of preventative measures against HIV infection. Interferon regulatory factor-1 (IRF1) plays a critical role both in host antiviral immunity and in HIV-1 replication. This study examined IRF1 expression regulation in the ex vivo peripheral blood mononuclear cells of HIV-exposed seronegative commercial sex workers who can be epidemiologically defined as relatively resistant to HIV infection (HIV-R), versus HIV-uninfected, susceptible controls (HIV-S). Whereas HIV-susceptible individuals demonstrated a biphasic, prolonged increase in IRF1 expression after interferon-γ stimulation, HIV-R individuals demonstrated a robust, but transient response. We also found that the IRF1 promoter in HIV-R was primed by increased basal histone deacetylase-2 binding, independently of transcription regulators, STAT1 and nuclear factor-κB/p65, implicating an epigenetic silencing mechanism. Interestingly, the transitory IRF1 response in HIV-R was sufficient in comparable regulation of interleukin-12 and interleukin-4 expression compared with the HIV-susceptible controls. This is the first study characterizing IRF1 responsiveness in individuals who demonstrate altered susceptibility to HIV infection. These data suggest that transitory IRF1 responsiveness in HIV-R may be one of the key contributors to the altered susceptibility to HIV infection during the early stages of primary HIV infection.
Introduction
Several studies of populations at high risk of HIV infection have identified individuals who are HIV-exposed but remain seronegative (HESN) and HIV-uninfected.1-4 Some groups have identified HESN groups that can be epidemiologically defined as relatively resistant to HIV infection (HIV-R).2,5 Understanding what protects these HIV-R individuals from acquiring HIV infection will help in developing preventative measures against infection. This study examined how the regulation of a key immunoregulatory factor, interferon regulatory factor-1 (IRF1) might differ in individuals who demonstrated altered susceptibility to HIV infection.
IRF1 belongs to the extensive IRF family of transcriptional activators and repressors. It is implicated in multiple biologic processes, including regulation of innate and adaptive immunity, cytokine signaling, apoptosis, and viral defense.6,7 IRF1, expressed at low levels in a variety of cell types, can be up regulated by type I and II interferon (IFN), as well as other cytokines, and viral infection (eg, HIV). Of particular interest, IRF1 is involved in HIV infection. The importance of IRF1 in activating the transcription of HIV genome during the early stage of HIV replication is demonstrated in Jurkat cells.8,9 Our group recently identified specific IRF1 polymorphisms that correlate with reduced susceptibility to HIV infection and reduced basal (by ∼ 60%) and IFN-γ-stimulated IRF1 protein expression in peripheral blood mononuclear cells (PBMCs).10 This suggests that reduction in IRF1 expression and responsiveness may contribute to altered susceptibility to HIV infection and reduced immune activation. As IRF1 seems to be such a key regulator and driver of HIV replication, we examined (1) IRF1 regulation at the mRNA and protein level in HIV-R individuals who lacked the previously reported “protective” genotypes and (2) IRF1 function in regulating the expression of immunologic genes.
Molecular regulation of IRF1 expression has been well defined in various cell lines and animal models.11 Several DNA elements in the IRF1 promoter proximal region are targets of various signaling pathways: γ-activated sequence (−110/−120), which binds STAT-1, as well as binding sites for nuclear factor-κB (NF-κB; −35/−45) and Sp1 (−200).7,12 Although data from such homogeneous in vitro systems are both instrumental and essential in defining the molecular mechanisms regulating IRF1 expression, how they relate to IRF1 regulation in directly ex vivo human cells and in the context of HIV susceptibility remains unclear. This study tested the hypothesis that IRF1 expression, its responsiveness to stimulation, and its molecular regulatory mechanisms in HIV-R women differ from that of control individuals susceptible to HIV infection (HIV-S).
The hypothesis was examined using whole PBMCs as IRF1 is expressed ubiquitously in all cell types and limited studies have detailed the immune cells primarily expressing IRF1. The HIV-R individuals are members of a well-characterized commercial sex-worker cohort with frequent exposure to virus but remain seronegative for more than 7 years. HIV-S individuals are new recruits of the same Kenyan cohort who have been in the commercial sex trade for less than 3 years. With constant exposure, even with strong counseling and prevention efforts (ie, condom use), our epidemiologic data suggest that approximately 85% of the HIV-S individuals in this cohort to succumb to infection pressure and eventually seroconvert. They are thus considered “epidemiologically susceptible” to HIV infection (HIV-S).5 We examined the kinetics of and molecular regulatory events in IRF1 responses to exogenous IFN-γ stimulation in ex vivo PBMCs of HIV-R and HIV-S individuals. By characterizing the binding of STAT1, NF-κB/p65, and histone deacetylase (HDAC) and the changes in histone (de)acetylation in regulating IRF1 responses in HIV-R and HIV-S individuals of our Kenyan sex-worker cohort, we sought a better understanding of the potential role of IRF1 regulation in altered susceptibility to HIV infection.
Herein, we report a strong but transient IRF1 response to exogenous IFN-γ in ex vivo PBMCs of HIV-R individuals. In comparison, robust, continuous responses were observed in HIV-S controls. Changes in IRF1 expression were correlated with the level of histone acetylation at the IRF1 promoter. These data suggest that the prompt initiation and silencing of IRF1 responses in HIV-R individuals may be an important driver for the resistant phenotype.
Methods
Study population
In total, 11 HIV-S and 18 HIV-R age-matched samples were chosen randomly from a well-characterized cohort of commercial sex workers from the Pumwani district of Nairobi, Kenya.13 The study was approved by the ethics review committees of the University of Manitoba and the Kenyatta National Hospital. Participants were assessed twice a year for follow-up. At each visit, blood samples were obtained for HIV-1 serologic analysis; demographic and behavioral data were also obtained. Participants were divided into 2 epidemiologically defined groups: HIV-R, if the individuals met our previously established definition of HESN (seronegative on enrollment, HIV-1 negative by serology and polymerase chain reaction [PCR] for > 7 years of follow-up); and HIV-S, if the individuals were enrolled within 2 years, in the sex trade for less than 3 years, and seronegative when the blood samples were taken.
Isolation of PBMCs and cell culture
PBMCs were isolated by Ficoll-Hypaque density gradient centrifugation, frozen in freezing solution (40% fetal bovine serum, 10% dimethyl sulfoxide, 50% RPMI culture media), in Mr Frosty freezing boxes (Nalgen) overnight at −80°C before transfer to liquid nitrogen for storage. Cryopreserved PBMCs were shipped in dry shippers to the University of Manitoba where experiments were performed. On thawing in 37°C water bath with constant agitation, PBMCs were immediately washed with 12 mL of culture media (cell viability > 90%) and cultured in RPMI 1640 (supplemented with 10% fetal bovine serum) at 37°C, 5% CO2 for 3 hours. The rested, unstimulated PBMCs were then cultured either alone in media or with IFN-γ (10 ng/mL, Sigma-Aldrich) for the time intervals indicated. At each time point, one-third of the cells were used for RNA isolation, and the other two-thirds of the cells were fixed with 1% formaldehyde for chromatin preparation.
Flow cytometry
PBMCs (3 × 105 cells per tube) were washed once with 2% fetal bovine serum in phosphate-buffered saline and then fixed with Perm/Fix Reagent (BD Biosciences), following the manufacturer's protocol. Cells were then washed twice with Perm/Wash Buffer and stained with primary antibody (specificity: IRF1, NF-κB/p65, rabbit Ig control). All primary antibodies were purchased from Abcam, with the exception of IRF1 (Santa Cruz Biotechnology). Fluorescein isothiocyanate–conjugated goat anti–rabbit secondary antibody (Jackson ImmunoResearch Laboratories) was used to visualize primary antibody binding. Stained cells were analyzed using BD FACSCalibur. All events were analyzed using CellQuest Pro Software Version 6.1.2 (BD Biosciences). Lymphocytes in PBMC samples were gated by including only single cells (forward scatter [FSC]-A vs FSC-H) and cells that fell within the lymphocyte gates (FSC-H vs side scatter [SSC-H]).
ChIP
Chromatin samples were prepared and used in immunoprecipitation assay as described previously with some modifications.14 Briefly, PBMCs (∼ 3-4 × 106) were fixed with 1% formaldehyde, lysed with NP-40 buffer, and then resuspended in 250 μL of sodium dodecyl sulfate nuclei-lysis buffer containing protease inhibitors (Roche Diagnostics) for sonication. Chromatin (200-500 bp in length) was precleared with protein A-Sepharose beads and immunoprecipitated with antibodies (specificity: STAT1, acetylated histone H4, NF-κB/p65, HDAC2, and rabbit Ig control). All antibodies were purchased from Abcam, with the exception of STAT1 and IRF1 antibodies (Santa Cruz Biotechnology). Eluted immune complexes were reverse cross-linked and treated with proteinase K to remove protein. Chromatin immunoprecipitation (ChIP)-DNA was then analyzed using quantitative PCR with specific target primer sets, synthesized by Invitrogen.
Quantitative RT-PCR
Total RNA was prepared as described previously, using Trizol (Sigma-Aldrich) and RNeasy MinElute Cleanup Kit (QIAGEN). RNA was treated with RNase-free DNase I before reverse transcription (RT; QIAGEN). Resulting cDNA was evaluated in quantitative PCR with primer sets specific for IRF1 primary transcripts (exon1-intron1), and mRNA (intron-less exon1-exon2 region) and I8S rRNA with proper RT controls. Annealing temperature for all primer sets was 60°C. All quantitative PCRs were performed with SYBR Green qPCR Master Mix (QIAGEN). Average threshold cycle (Ct) from duplicated wells (with coefficient of variation < 10%) was used to calculate the relative change in IRF1 expression, using 18S rRNA for normalization and the media alone culture condition as a reference (ΔΔCt program, Applied Biosystems). Products from ChIP were analyzed with primer sets specific for IRF1 proximal promoter, and intron7 (annealing temperature, 60°C). Fold changes in transcription factor binding or histone acetylation were calculated using the input DNA for normalization and the media alone culture condition as a reference (ΔΔCt program; Applied Biosystems).
Data analysis
Data from quantitative PCR were analyzed using ABI 7500 System Sequence Detection Software, Version 1.40.25. Statistical analyses were performed with GraphPad Prism, Version 4.0. Unpaired t test was used to determine whether median values differed significantly.
Results
Basal IRF1 RNA and protein expression in HIV-R individuals was compared with HIV-S controls
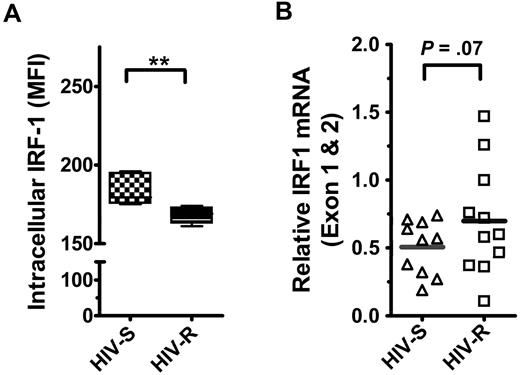
We examined the basal expression and regulation of IRF1 in human PBMCs from HIV-S and HIV-R individuals to test the hypothesis that HIV-R individuals had reduced level of basal IRF1 expression. IRF1 protein and RNA expression in ex vivo unstimulated PBMCs were measured using intracellular flow cytometry and quantitative RT-PCR, respectively. All lymphocytes in the PBMC populations (ie, HIV-R and HIV-S) examined were stained positive for intracellular IRF1. We found that the level of IRF1 protein expression in all leukocytes examined was slightly reduced (∼ 8%) in the HIV-R individuals (n = 11, mean fluorescence intensity = 168), compared with HIV-S controls (n = 10, mean fluorescence intensity = 183, P = .006, Figure 1A). However, interestingly, we noticed a trend toward an increase in basal IRF1 mRNA expression in the HIV-R individuals (P = .07, Figure 1B). This observation suggests that, unlike our earlier study,10 these HIV-R individuals who lack the protective IRF1 genotype do not have marked reduction in IRF1 basal expression.
The expression and epigenetic regulation of IRF1 gene inex vivounstimulated PBMCs from HIV-S and HIV-R individuals. (A) Intracellular IRF1 protein expression in ex vivo PBMCs from HIV-S and HIV-R (n = 6, each) participants was measured using flow cytometric analyses. Intracellular staining was performed using IRF1 specific antibody and fluorescein isothiocyanate-labeled goat antirabbit antibody. Background signal from the secondary antibody alone was within the first log (100-101). (B) Intracellular IRF1 mRNA expression in ex vivo PBMCs from HIV-S (▵, n = 10) and HIV-R (□, n = 11) participants was assessed using quantitative RT-PCR. Bars represent mean values. **P < .005. Not significant (P > .05).
The expression and epigenetic regulation of IRF1 gene inex vivounstimulated PBMCs from HIV-S and HIV-R individuals. (A) Intracellular IRF1 protein expression in ex vivo PBMCs from HIV-S and HIV-R (n = 6, each) participants was measured using flow cytometric analyses. Intracellular staining was performed using IRF1 specific antibody and fluorescein isothiocyanate-labeled goat antirabbit antibody. Background signal from the secondary antibody alone was within the first log (100-101). (B) Intracellular IRF1 mRNA expression in ex vivo PBMCs from HIV-S (▵, n = 10) and HIV-R (□, n = 11) participants was assessed using quantitative RT-PCR. Bars represent mean values. **P < .005. Not significant (P > .05).
HIV-R individuals have slightly elevated histone H4 acetylation but similar STAT1 and NF-κB/p65 binding at the IRF1 promoter
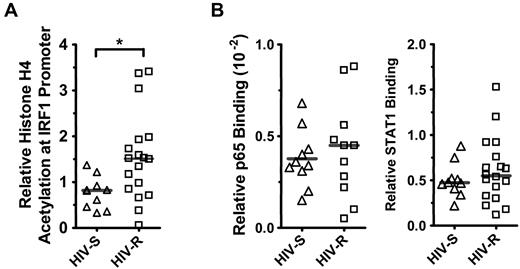
It is well established that epigenetic mechanisms govern the expression potential of a gene,15-17 and previous data have demonstrated that the acetylation level of histones at the IRF1 promoter correlated with IRF1 expression in Nb2 T lymphoma cells.18 Here, we examined whether the IRF1 expression potential in HIV-R individuals differs from that of the HIV-S controls by measuring the basal levels of histone acetylation and the binding of the transcription regulators STAT1 and NF-κB/p65 at the IRF1 promoter in primary PBMCs from HIV-S (n = 10) and HIV-R (n = 18) individuals. ChIP and quantitative PCR were used to assess acetylated histone H4, and we further examined the binding of the transcription factors STAT1 and NF-κB/p65 at IRF1 gene loci in ex vivo, unstimulated PBMCs (Figure 2A). Supporting the observed trend to an increase in basal IRF1 mRNA in HIV-R individuals, approximately 2-fold increase in acetylated histone H4 at the IRF1 promoter was observed in HIV-R individuals, versus that in the HIV-S controls (Figure 2A; P = .01). However, there were no differences in the basal STAT1 or NF-κB/p65 binding between the HIV-R and HIV-S groups (Figure 2B), suggesting that, although HIV-R individuals may have a slightly increased capacity for transcription because of increased histone H4 acetylation, equivalent binding of the transcription factors STAT1 and NF-κB/p65 between 2 groups indicates a similar IRF1 expression potential.
Histone H4 acetylation and NF-κB/p65 and STAT1 binding at IRF1 promoter inex vivounstimulated PBMCs. (A) Basal histone H4 acetylation at IRF1 locus was assessed in the ex vivo PBMCs from HIV-S (▵, n = 10) and HIV-R (□, n = 18) individuals using ChIP assay and analyzed with quantitative PCR. (B) NF-κB/p65 binding to IRF1 promoter in ex vivo PBMCs of HIV-S (▵, n = 10) and HIV-R (□, n = 11) participants was examined using ChIP and analyzed with quantitative PCR. Basal level of STAT1 binding to IRF1 promoter in the ex vivo, unstimulated PBMCs of HIV-S (▵, n = 10) and HIV-R (□, n = 18) individuals was also examined. Bars represent mean values. *P < .05. Not significant (P > .05).
Histone H4 acetylation and NF-κB/p65 and STAT1 binding at IRF1 promoter inex vivounstimulated PBMCs. (A) Basal histone H4 acetylation at IRF1 locus was assessed in the ex vivo PBMCs from HIV-S (▵, n = 10) and HIV-R (□, n = 18) individuals using ChIP assay and analyzed with quantitative PCR. (B) NF-κB/p65 binding to IRF1 promoter in ex vivo PBMCs of HIV-S (▵, n = 10) and HIV-R (□, n = 11) participants was examined using ChIP and analyzed with quantitative PCR. Basal level of STAT1 binding to IRF1 promoter in the ex vivo, unstimulated PBMCs of HIV-S (▵, n = 10) and HIV-R (□, n = 18) individuals was also examined. Bars represent mean values. *P < .05. Not significant (P > .05).
IRF1 responses to IFN-γ stimulation are maintained in HIV-S controls but rapidly down regulated in HIV-R individuals
IFN-γ production is a key component of the innate and adaptive immune response to viral infections, including HIV,19-21 and is an effective inducer of IRF1.19 Previous data from our group have demonstrated that polymorphisms in the IRF1 gene that associate with protection from HIV infection also associate with reduced responsiveness to IFN-γ stimulation.10 Here, we examined whether IRF1 response to IFN-γ was also decreased in HIV-R women who do not have the “protective” IRF1 genotype (Figure 3). Ex vivo, unstimulated PBMCs from HIV-R (n = 12) and HIV-S (n = 10) individuals were exposed to exogenous IFN-γ in culture for the indicated lengths of time. The kinetics of IRF1 primary transcripts (Figure 3A), mRNA level (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article), and protein expression (Figure 3B-C) were studied using quantitative RT-PCR and flow cytometry, respectively. Cells cultured in media without IFN-γ for the same indicated length of time served as unstimulated controls. Culture media alone had no effect on cellular viability, IRF1, interleukin-4 (IL-4), and IL-12p35 transcription, 18s rRNA level, or IRF1 protein expression (data not shown).
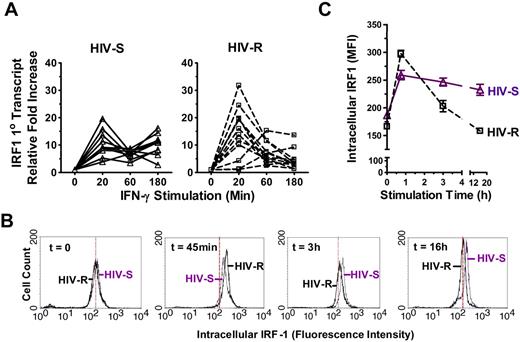
Kinetics of IRF1 responses to exogenous IFN-γ stimulation. (A) Ex vivo PBMCs from HIV-S (▵, n = 10) and HIV-R (□, n = 12) individuals were stimulated with exogenous IFN-γ (10 ng/mL). At the indicated time points, RNA were isolated and IRF1 primary RNA transcripts levels were examined using quantitative RT-PCR. There was no significant difference in transcript levels between the unstimulated samples (time = 0, 20, 60, or 180 minutes). Unstimulated sample from time = 0 is used as reference for calculating relative fold increases. The RNA transcripts were normalized to endogenous 18S RNA. (B-C) Intracellular expression of IRF1 in ex vivo PBMCs from 6 HIV-S and 6 HIV-R individuals was examined at 45 minutes, 3 hours, and 16 hours after IFN-γ (10 ng/mL) stimulation. Intracellular staining was performed using IRF1 specific antibody and fluorescein isothiocyanate-labeled goat antirabbit antibody. Background signal from the secondary antibody alone was within the first log (100-101). The histogram plots of different time points were representative of the individuals examined in each group. Compilation of data from 6 independent samples from each group was graphed in panel C. The y-axis graphed the mean fluorescence intensity (MFI) of IRF1 staining.
Kinetics of IRF1 responses to exogenous IFN-γ stimulation. (A) Ex vivo PBMCs from HIV-S (▵, n = 10) and HIV-R (□, n = 12) individuals were stimulated with exogenous IFN-γ (10 ng/mL). At the indicated time points, RNA were isolated and IRF1 primary RNA transcripts levels were examined using quantitative RT-PCR. There was no significant difference in transcript levels between the unstimulated samples (time = 0, 20, 60, or 180 minutes). Unstimulated sample from time = 0 is used as reference for calculating relative fold increases. The RNA transcripts were normalized to endogenous 18S RNA. (B-C) Intracellular expression of IRF1 in ex vivo PBMCs from 6 HIV-S and 6 HIV-R individuals was examined at 45 minutes, 3 hours, and 16 hours after IFN-γ (10 ng/mL) stimulation. Intracellular staining was performed using IRF1 specific antibody and fluorescein isothiocyanate-labeled goat antirabbit antibody. Background signal from the secondary antibody alone was within the first log (100-101). The histogram plots of different time points were representative of the individuals examined in each group. Compilation of data from 6 independent samples from each group was graphed in panel C. The y-axis graphed the mean fluorescence intensity (MFI) of IRF1 staining.
A similar rapid and strong increase in IRF1 primary transcripts was detected in both HIV-S and HIV-R groups compared with unstimulated controls (P = .0004 and .003, respectively), within 20 minutes of IFN-γ exposure. Primary transcripts (newly synthesized, unspliced RNA) are a direct readout of transcriptional activation. At 60 minutes after stimulation, IRF1 primary transcripts declined abruptly in both study groups and remained low in HIV-R individuals (see Figure 7C, up to 16 hours) but subsequently increased in the HIV-S individuals, demonstrating a biphasic response to IFN-γ stimulation in HIV-S, versus an apparent silencing of IRF1 transcription in the HIV-R group (P < .02). The sustained biphasic IRF response in HIV-S was observed up to 16 hours after stimulation (data not shown). The kinetics of IRF1 responsiveness was also reflected in IRF1 protein levels with delayed kinetics (Figure 3B-C). On exposure to exogenous IFN-γ, the intracellular IRF1 protein level increased rapidly within 45 minutes in both HIV-R and HIV-S individuals (n = 6 in each group). Mirroring the changes observed in the primary mRNA transcripts, a sharp decline in IRF1 protein level was observed in HIV-R individuals at 3 hours after stimulation (Figure 3C). In contrast, the intracellular IRF1 protein expression reached a plateau at 3 hours after stimulation in the HIV-S controls. The intracellular IRF1 protein expression in all leukocytes examined was similarly reduced in the HIV-R individuals (Figure 3B; time = 3 hours, 16 hours, ie, one single peak). Together, the data illustrate a sustained, biphasic IRF1 response to exogenous IFN-γ stimulation in HIV-S individuals, but a transitory response in HIV-R individuals.
Increased IRF1 recruitment to IRF1 target genes (ie, IL-4 and IL-12) after IFN-γ stimulation is found in both HIV-S and HIV-R groups
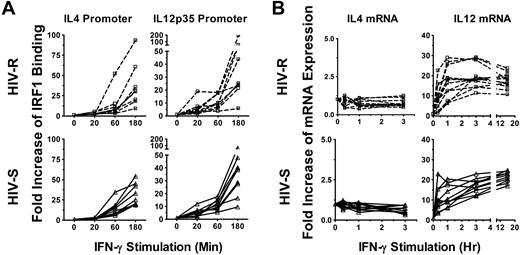
The expression of IL-12 or IL-4 is one of the key signals in the initiation of an adaptive cellular (IL-12) or humoral (IL-4) immune response. IRF1 has been shown to bind to both the IL-12p35 subunit and IL-4 promoters, up-regulating IL-12 and repressing IL-4, and is a critical regulator of the function and differentiation of adaptive immunity. Here, we tested the hypothesis that the rapid, transient increases in IRF1 expression was sufficient to activate and/or silence IRF1 target genes responsible in part for inducing adaptive immunity in HIV-R individuals. Binding of IRF1 to IL-4 and IL-12p35 promoters in ex vivo PBMCs, after exogenous IFN-γ stimulation was assessed using ChIP-quantitative PCR analysis (Figure 4A). IRF1 recruitment to IL-4 and IL-12p35 promoters was significantly increased at 60 minutes after stimulation and was comparable in both HIV-R and HIV-S individuals (Figure 4A). Furthermore, IRF1 recruitment continued to increase 3 hours after stimulation, providing evidence that the transient increase in IRF1 expression is sufficient for IRF1 function in regulating IL-4 and IL-12p35 genes in PBMCs.
Recruitment of IRF1 to IL-12p35 and IL-4 promoters and their mRNA expression after exogenous IFN-γ stimulation. (A) Ex vivo PBMCs from HIV-S (n = 9, ▵) and HIV-R (n = 9, □) individuals were stimulated with exogenous IFN-γ (10 ng/mL). At the indicated time points, chromatin was isolated and immunoprecipitated with antibodies specific for IRF1 chromatin-immunoprecipitated DNA products were analyzed for the presence of IL-12p35 and IL-4 promoter using quantitative PCR. Quantitative PCR signals were normalized to input DNA. There was no difference in the levels of IRF1 binding between the unstimulated samples, cultured in media alone for 0, 60, or 180 minutes. Unstimulated sample from time = 0 is used as reference for calculating relative fold increases. (B) RNA were also isolated from the PBMCs of HIV-S (n = 10, ▵) and HIV-R (n = 11, □) individuals, after IFN-γ stimulation. IL-12p35 and IL-4 mRNA levels were examined using quantitative RT-PCR. There was no significant difference in mRNA levels between the unstimulated samples (time = 0, 20, 60, or 180 minutes). Unstimulated sample from time = 0 is used as reference for calculating relative fold increases. The RNA transcripts were normalized to endogenous 18S rRNA.
Recruitment of IRF1 to IL-12p35 and IL-4 promoters and their mRNA expression after exogenous IFN-γ stimulation. (A) Ex vivo PBMCs from HIV-S (n = 9, ▵) and HIV-R (n = 9, □) individuals were stimulated with exogenous IFN-γ (10 ng/mL). At the indicated time points, chromatin was isolated and immunoprecipitated with antibodies specific for IRF1 chromatin-immunoprecipitated DNA products were analyzed for the presence of IL-12p35 and IL-4 promoter using quantitative PCR. Quantitative PCR signals were normalized to input DNA. There was no difference in the levels of IRF1 binding between the unstimulated samples, cultured in media alone for 0, 60, or 180 minutes. Unstimulated sample from time = 0 is used as reference for calculating relative fold increases. (B) RNA were also isolated from the PBMCs of HIV-S (n = 10, ▵) and HIV-R (n = 11, □) individuals, after IFN-γ stimulation. IL-12p35 and IL-4 mRNA levels were examined using quantitative RT-PCR. There was no significant difference in mRNA levels between the unstimulated samples (time = 0, 20, 60, or 180 minutes). Unstimulated sample from time = 0 is used as reference for calculating relative fold increases. The RNA transcripts were normalized to endogenous 18S rRNA.
We next examined the mRNA levels of IL-4 and IL-12p35 using quantitative RT-PCR (Figure 4B). IRF1 was previously reported to transactivate IL-12p35 transcription while repressing IL-4 transcription, hence promoting Th1-like, or cellular immune responses. Here, we observed increases in IL-12 mRNA and the maintenance of low IL-4 mRNA expression, after IFN-γ stimulation in both HIV-R and HIV-S individuals, supporting the notion that transient IRF1 responses in HIV-R play a functional role in regulating immunologic gene expression.
IFN-γ induced IRF1 expression closely correlates with the level of acetylated histone H4 at the IRF1 gene loci
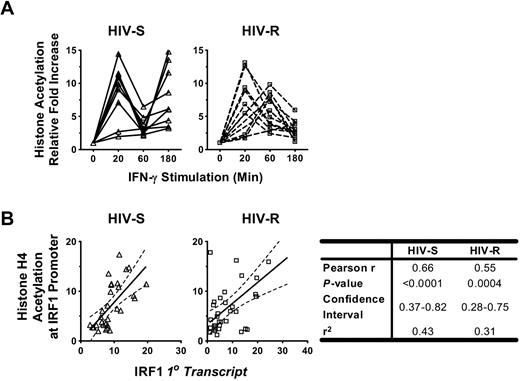
We investigated whether histone acetylation at the IRF1 locus was altered during IFN-γ stimulation and how histone acetylation might be related to the dynamic changes of IRF1 expression in PBMCs from HIV-R individuals. Changes in histone H4 acetylation, after exogenous IFN-γ stimulation, were studied using ChIP-quantitative PCR and primers specific for IRF1 promoter (Figure 5A) and intron7 (supplemental Figure 2). The IRF1 intron 7 was examined to determine whether the changes in epigenetic modifications would spread across the majority of the gene locus. Mirroring the changes in IRF1 primary transcripts (Figure 3A), rapid, robust increases in histone acetylation, followed by an immediate reduction, were observed in both HIV-R (n = 11) and HIV-S (n = 10) individuals (Figure 5A). Whereas the HIV-R individuals maintained low levels of acetylation for more than 60 minutes after stimulation, similar to the RNA expression data, the HIV-S controls again showed a biphasic response with increased histone acetylation at the promoter. In both groups, the pattern of changes in acetylated histone H4 spread to the IRF1 intron7 region (supplemental Figure 2). As expected, the pattern of histone H4 acetylation at the IRF1 promoter was remarkably similar to the kinetics of the IRF1 primary transcript (Figure 3A) and mRNA levels (supplemental Figure 1) and correlated strongly with the level of IRF1 primary transcripts in both groups, with Pearson r values ranging from 0.55 to 0.66 (P ≤ .0004; Figure 5B) when statistically analyzed. The tight association between histone acetylation level and transcriptional activity suggests the importance of epigenetic mechanisms in regulating IRF1 responses to IFN-γ.
Changes in histone H4 acetylation level at IRF1 promoter, their relation with the level of IRF1 transcripts, and HDAC2 recruitments after IFN-γ stimulation. (A) Ex vivo PBMCs from HIV-S (n = 10, ▵) and HIV-R (n = 11, □) individuals were stimulated with exogenous IFN-γ (10 ng/mL). At the indicated time points, chromatin was isolated and immunoprecipitated with antibodies specific for acetylated histone H4. Chromatin-immunoprecipitated DNA products were analyzed for the presence of IRF1 promoter using quantitative PCR. Quantitative PCR signals were normalized to input DNA. There was no difference in the level of acetylated histone H4 between the unstimulated samples cultured in media alone for 0, 60, or 180 minutes. Unstimulated sample from time = 0 is used as reference for calculating relative fold increases. (B) The correlation between the IRF1 primary transcripts level (from Figure 2A) and the level of histone H4 acetylation at IRF1 promoter (from Figure 4A) in each study group was analyzed using Pearson correlation test. The results of linear regression were graphed with 95% confidence interval for each group. The results of Pearson correlation analyses were tabulated.
Changes in histone H4 acetylation level at IRF1 promoter, their relation with the level of IRF1 transcripts, and HDAC2 recruitments after IFN-γ stimulation. (A) Ex vivo PBMCs from HIV-S (n = 10, ▵) and HIV-R (n = 11, □) individuals were stimulated with exogenous IFN-γ (10 ng/mL). At the indicated time points, chromatin was isolated and immunoprecipitated with antibodies specific for acetylated histone H4. Chromatin-immunoprecipitated DNA products were analyzed for the presence of IRF1 promoter using quantitative PCR. Quantitative PCR signals were normalized to input DNA. There was no difference in the level of acetylated histone H4 between the unstimulated samples cultured in media alone for 0, 60, or 180 minutes. Unstimulated sample from time = 0 is used as reference for calculating relative fold increases. (B) The correlation between the IRF1 primary transcripts level (from Figure 2A) and the level of histone H4 acetylation at IRF1 promoter (from Figure 4A) in each study group was analyzed using Pearson correlation test. The results of linear regression were graphed with 95% confidence interval for each group. The results of Pearson correlation analyses were tabulated.
The kinetics of IFN-γ induced recruitment of STAT1 and NF-κB/p65 to the IRF1 promoter mirrors that of IRF1 expression and promoter acetylation
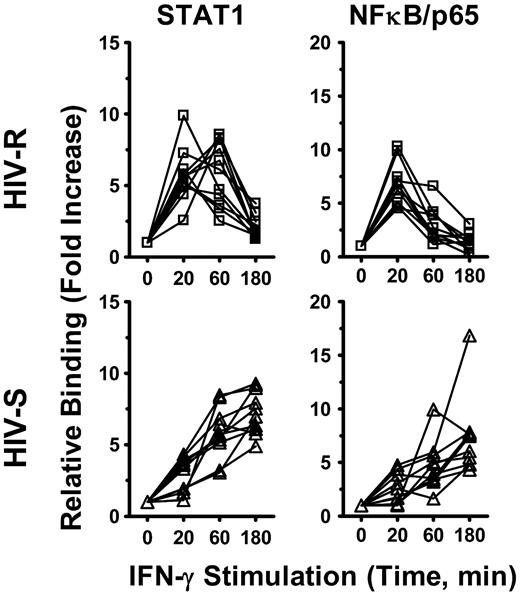
Both STAT1 and NF-κB binding has previously been shown to enhance the histone acetylation of targeted genes.22-24 We investigated whether the recruitment of STAT1 and NF-κB/p65 transcription factor to IRF1 promoter had a role in IFN-γ-induced IRF1 responses and the different kinetics of expression observed between the HIV-R and HIV-S individuals, using ChIP-quantitative PCR. Binding of these transcription factors to IRF1 intron7 region was used as background binding in calculation. After IFN-γ stimulation, both STAT1 and NF-κB/p65 were recruited to the IRF1 promoter of PBMCs from both HIV-R and HIV-S and rapidly decreased after 60 minutes in the HIV-R individuals but remained robust and sustained in the HIV-S controls (Figure 6). The kinetics of STAT1 and NF-κB/p65 recruitment mirrored that of IRF1 mRNA level (Figure 3A) and histone H4 acetylation at IRF1 promoter in HIV-R and HIV-S (Figure 2B), suggesting that both STAT1 and NF-κB/p65 are involved in regulating IRF1 activation in primary PBMCs.
NF-κB/p65 and STAT1 recruitment to IRF1 promoter after exogenous IFN-γ stimulation. Ex vivo PBMCs from HIV-S (n = 10, ▵) and HIV-R (n = 11, □) individuals were stimulated with exogenous IFN-γ (10 ng/mL). At the indicated time points after stimulation, chromatin was isolated and immunoprecipitated with antibodies specific for STAT1 and NF-κB/p65. Chromatin-immunoprecipitated DNA products were analyzed for the presence of IRF1 promoter using quantitative PCR. Quantitative PCR signals were normalized to input DNA. There was no difference in the levels of STAT1 or NF-κB/p65 between the unstimulated samples cultured in media alone for 0, 60, or 180 minutes. Unstimulated sample from time = 0 is used as reference for calculating relative fold increases.
NF-κB/p65 and STAT1 recruitment to IRF1 promoter after exogenous IFN-γ stimulation. Ex vivo PBMCs from HIV-S (n = 10, ▵) and HIV-R (n = 11, □) individuals were stimulated with exogenous IFN-γ (10 ng/mL). At the indicated time points after stimulation, chromatin was isolated and immunoprecipitated with antibodies specific for STAT1 and NF-κB/p65. Chromatin-immunoprecipitated DNA products were analyzed for the presence of IRF1 promoter using quantitative PCR. Quantitative PCR signals were normalized to input DNA. There was no difference in the levels of STAT1 or NF-κB/p65 between the unstimulated samples cultured in media alone for 0, 60, or 180 minutes. Unstimulated sample from time = 0 is used as reference for calculating relative fold increases.
HIV-R individuals have higher basal level of HDAC2 binding at IRF1 promoter
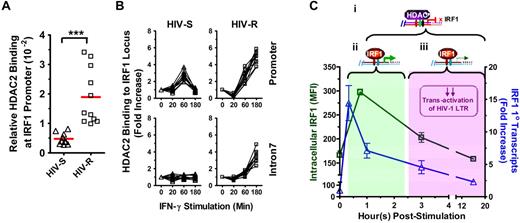
Epigenetic regulators of histone acetylation are histone acetyltransferase and HDACs, which remove the acetyl modification. To determine whether the epigenetic regulation of histone deacetylation might be involved in the rapid silencing of IRF1 response, we examined whether the basal level of HDAC binding at the IRF1 promoter differs between the HIV-R and HIV-S individuals (Figure 7A). HDAC1, HDAC2, and HDAC3 are closely related and have been implicated in the regulation of histone acetylation, and transactivation of NF-κB and STAT1.25-27 Using ChIP and quantitative PCR analyses, we found HDAC2, but not HDAC1 or HDAC3 (data not shown), binding to the IRF1 promoter in the ex vivo PBMCs of HIV-R (n = 10) and HIV-S (n = 11) individuals (Figure 7A). Four-fold more HDAC2 binding to the IRF1 promoter was detected in the ex vivo PBMCs samples from HIV-R, compared with HIV-S controls (P = .0002), suggesting that the IRF1 promoter in HIV-R individuals may be “primed” for silencing before IFN-γ stimulation.
Basal binding and recruitment of HDAC2 to IRF1 gene locus. (A) Basal HDAC2 binding at IRF1 promoter was assessed in the ex vivo PBMCs from HIV-S (▵, n = 10) and HIV-R (□, n = 11) individuals using ChIP-quantitative PCR with antibodies specific for HDAC2. Bars represent mean values. ***P < .0005. Not significant (P > .05). (B) HDAC2 recruitment to IRF1 promoter and intron7 after exogenous IFN-γ stimulation was examined at the indicated time points after IFN-γ stimulation. Quantitative PCR signals were normalized to input DNA. There was no difference in the levels of HDAC2 between the unstimulated samples cultured in media alone for 0, 60, or 180 minutes. Unstimulated sample from time = 0 is used as reference for calculating relative fold increases. (C) Study model: IRF1-responsive potential in HIV-R commercial sex workers and its implication in resistance to HIV acquisition. On stimulation by IFN-γ, IRF1 expression is robustly increased within 20 minutes at the transcriptional level. (i) Increased IRF1 binding to its target genes (eg, IL-12p35, IL-4) and the regulation of target gene express are also observed shortly after stimulation. (ii) Within an hour after stimulation, increases in IRF1 expression are controlled by the recruitment of HDAC2 to IRF1 loci and histone deacetylation spreading across the IRF1 gene. The transitory increase in IRF1 expression is sufficient in inducing comparable immune responses. (iii) At the same time, the rapid silencing of IRF1 expression may have a role in the HIV resistance by curtailing the transactivation of HIV-1 LTR during the early stages of viral infection and hence, allowing time for innate and acquired immunity of develop.
Basal binding and recruitment of HDAC2 to IRF1 gene locus. (A) Basal HDAC2 binding at IRF1 promoter was assessed in the ex vivo PBMCs from HIV-S (▵, n = 10) and HIV-R (□, n = 11) individuals using ChIP-quantitative PCR with antibodies specific for HDAC2. Bars represent mean values. ***P < .0005. Not significant (P > .05). (B) HDAC2 recruitment to IRF1 promoter and intron7 after exogenous IFN-γ stimulation was examined at the indicated time points after IFN-γ stimulation. Quantitative PCR signals were normalized to input DNA. There was no difference in the levels of HDAC2 between the unstimulated samples cultured in media alone for 0, 60, or 180 minutes. Unstimulated sample from time = 0 is used as reference for calculating relative fold increases. (C) Study model: IRF1-responsive potential in HIV-R commercial sex workers and its implication in resistance to HIV acquisition. On stimulation by IFN-γ, IRF1 expression is robustly increased within 20 minutes at the transcriptional level. (i) Increased IRF1 binding to its target genes (eg, IL-12p35, IL-4) and the regulation of target gene express are also observed shortly after stimulation. (ii) Within an hour after stimulation, increases in IRF1 expression are controlled by the recruitment of HDAC2 to IRF1 loci and histone deacetylation spreading across the IRF1 gene. The transitory increase in IRF1 expression is sufficient in inducing comparable immune responses. (iii) At the same time, the rapid silencing of IRF1 expression may have a role in the HIV resistance by curtailing the transactivation of HIV-1 LTR during the early stages of viral infection and hence, allowing time for innate and acquired immunity of develop.
Rapid silencing of the IRF1 response involves recruitment of HDAC2 to the IRF1 locus and the loss of histone acetylation throughout IRF locus
To further examine whether HDAC2 was involved in the changes of IRF1 gene expression and histone H4 acetylation at IRF1 locus after exogenous IFN-γ stimulation, the kinetics of HDAC2 recruitment to IRF1 promoter and intron7 was studied using ChIP-quantitative PCR (Figure 7B). IRF1 intron7 was examined to determine whether HDAC2 binding could spread across the IRF1 gene locus, as the histone deacetylation did previously. Significant increases in HDAC2 binding to the IRF1 promoter were observed in the PBMCs of HIV-S (n = 10) and HIV-R individuals (n = 11), at 60 minutes after IFN-γ stimulation (Figure 7B). Whereas HDAC2 recruitment to the IRF1 promoter and intron7 region continued to increase over time in HIV-R samples, HDAC2 binding to IRF1 promoter in HIV-S individuals was reduced to basal level at 180 minutes after stimulation. Thus, HDAC2 recruitment precisely paralleled the deacetylation (Figure 5A) and production of primary transcripts (Figure 3A). Similarly, the absence of HDAC2 binding to IRF1 intron7 coincided with the minimum deacetylation of histone H4 at intron7 in the HIV-S individuals (Figure 7B). Taken together, the data suggest that recruitment of HDAC2 and spreading of epigenetic modification across the IRF1 gene locus14 are critical for regulating IRF1 responsiveness. Further, these data together reveal that the regulation of IRF1 responsiveness is significantly altered in HIV-R individualscompared with susceptible controls and that inducing and maintaining IRF1 silencing may play a role in mediating resistance to infection by HIV-1.
Discussion
A transitory, robust IRF1 response in HIV-R individuals and its implication in HIV acquisition
In vitro HIV infection has been shown to increase IRF1 expression on virus entry, both in Jurkat T-cell lines and in primary CD4+ T cells, before the expression of HIV Tat protein.28 The significance of such increase was demonstrated in Jurkat T cells, stably expressing IRF1-specific siRNA. Sgarbanti et al showed that reducing IRF1 expression resulted in significantly decreased transactivation of the HIV long terminal repeat (LTR) and, thus, viral gene expression.9 However, it is not known whether altered IRF1 expression plays any role in vivo in individuals exhibiting reduced susceptibility to HIV acquisition (ie, HIV-R). In our earlier studies, we were able to demonstrate that genetic polymorphisms in IRF1 associate with the HIV resistance phenotype and further associate with reduced IRF1 expression and responsiveness to exogenous IFN-γ stimulation.10 In a more recent study, we showed that cells from individuals with reduced IRF1 expression have a reduced capacity to transactivate the HIV-1 LTR, when the cells were infected with a single-cycle HIV-1-VSVg pseudovirus construct expressing a luciferase reporter gene insert.29 These findings collectively demonstrate a strong relationship between reduced IRF1 expression and a reduced susceptibility to HIV. Nevertheless, not all HIV-R individuals have the “protective” IRF1 genotype. Data from this study demonstrate that HIV-R individuals who lack the “protective” IRF1 genotype exhibit reduced IRF1 responsive potential (Figure 3), albeit without a significant decrease in basal levels of IRF1 expression (Figure 1), further strengthening the relationship between control of IRF1 expression and the susceptibility to HIV infection.
Yet it is not clear from these genetic and in vitro studies9,10,28-30 how reduced IRF1 expression and responses could reduce the susceptibility to viral infection and, at the same time, be capable of generating an adaptive antiviral immune response.31-35 IRF1 is an important antiviral molecule and probably important in HIV susceptibility and pathogenesis.6,19,36 After simian immunodeficiency virus (SIV) infection, IFN-(α,β)-producing plasmatoid dendritic cell numbers37 and large increases in IFN-γ expression were found in cervical vaginal tissues.38 This is especially interesting as IRF1 is a critical transcriptional regulator of IFN-(α,β,γ) signaling. In addition, during pathogenic SIV infection, rhesus macaques who demonstrate SIV-induced immunopathology failed to up-regulate IRF1 expression during the early stage of infection,39,40 suggesting that increased IRF1 expression during early stage of SIV infection may be critical in triggering host responses capable of controlling SIV-induced pathogenesis. Genes that were uniquely increased in expression (eg, IDO/INDO, CD274/B7-H1) in the nonpathogenic phenotype and their protein have been reported to be immunosuppressive and are targets of IRF1.41-43 Although these observations support a role for IRF1 in the immunopathology of HIV, this raises the issue that altered IRF1 regulatory processes that may protect against infection (such as the ones presented in our study) may be different from the processes that regulate protection against HIV/SIV pathogenesis observed in the macaque studies. Collectively, these studies support the need for further studies to define the perhaps dichotomous role of IRF1 regulation in SIV/HIV disease pathogenesis and altered susceptibility to infection.
Our study is the first to define the early kinetics of IRF1 response to IFN-γ stimulation and the implied regulatory mechanisms in PBMCs from HIV-R and HIV-S individuals. We found that both HIV-R and HIV-S individuals had robust IRF1 response to exogenous IFN-γ stimulation, but it was transient and immediately controlled in the HIV-R individuals (Figure 3A-B). In contrast, the PBMCs from susceptible controls showed a biphasic response and failed to silence IRF1 responses (Figure 3). This suggests that (1) both groups of individuals can potentially induce IRF1-mediated immune responses and (2) the timely silencing of IRF1 response may be critical during the early stage of infection and perhaps has a functional role in altered susceptibility to HIV infection (Figure 7C).
This study examined IRF1 expression and responses in whole PBMCs because both IRF16 and IFN-γ receptor 1 are expressed in all blood leukocytes and myeloid cells44 (www.symatlas.gnf.org), and limited studies have detailed its expression in specific leukocyte subsets and their role in resistance to HIV infection. Studies are now underway to examine IRF1 regulation in leukocyte subsets; these studies should yield more insights in the role of IRF1 in resistance to HIV infection and will probably provide more relevant information as to the mechanisms behind IRF1's role in altered susceptibility to HIV infection.
Transient IRF1 expression in HIV-R individuals is sufficient in regulating cytokine gene expression
IRF1 is a crucial regulator in the development and function of adaptive immunity. Mice deficient in IRF1 fail to produce IL-12 and IL-15 and have a reduced number of mature CD8+ T cells.45,46 This study found that the transient IRF1 response in HIV-R individuals was sufficient in inducing comparable IRF1 binding to IL-12p35 and IL-4 promoters as in HIV-S individuals (Figure 4). And, as expected, IL-12 mRNA expression was increased while suppressed IL-4 expression was maintained, up to 16 hours after stimulation. However, the comparable binding between groups to the cytokine promoter(s) suggests that sufficient IRF1 protein is available in both groups to bind to HIV-1 LTR and possibly drive HIV replication. However, the critical difference between study groups was that, in those with demonstrable altered susceptibility to HIV-1, the IRF1 response was transient and rapidly repressed, suggesting that perhaps IRF1 binding to HIV-1 LTR and thus HIV-1 replication could be curtailed, allowing time for innate and acquired immunity to develop. We are currently studying the effects of reduced IRF1 expression on the transactivation of HIV-1 LTR in ex vivo CD3+ T cells (from normal healthy blood donors), using siRNA approaches. Preliminary data suggest that a 20% to 30% reduction in IRF1 expression at the time of infection was sufficient in blocking HIV-1 LTR transcription (R.-C.S., X.-J. Yao, J.K., W.J., F.A.P., and T.B.B., manuscript in preparation), without affecting the regulation of IL-12 and IL-4 mRNA expression. Taken together, the robust, but transient, IRF1-responsive potential in HIV-R individuals may be one of the keys to initiate antiviral responses with a minimum effect on viral replication during the early stage of primary HIV infection. Clearly, further prospective studies need to be conducted to determine the causality of this association of altered IRF1 expression and resistance to infection by HIV. It is intriguing to consider what the fate of the single subject within the “susceptible” group who demonstrated IRF1 responsiveness patterns compared with the HIV-R subjects. It begs the question: will they go on to become HIV-resistant? Our epidemiologic studies demonstrate that 10% of initially HIV-uninfected subjects will eventually meet this definition of HIV-R. Further prospective studies are underway to examine these intriguing possibilities.
Epigenetic regulation of the timely silencing of IRF1 expression in HIV-R individuals
This study identified the regulatory role of epigenetic controls, as characterized by histone acetylation and recruitments of HDAC2 in the IRF1 response to IFN-γ (Figures 5, 7). The level of histone acetylation at IRF1 promoter correlates strongly with the level of IRF1 primary transcripts in both HIV-R and HIV-S groups (Figure 5B), suggesting that the IRF1 responsive potential is regulated by epigenetic controls. Of particular interest, we found approximately 4-fold higher HDAC2 binding to IRF1 promoter in the ex vivo unstimulated PBMCs of HIV-R versus the HIV-S controls (Figure 7A). In contrast, the basal binding of other transcription regulators examined (eg, STAT1 and NF-κB/p65) was clearly not different between the 2 groups (Figure 2B). The fact that the ex vivo HDAC2 binding in HIV-R does not agree with the ex vivo IRF1 mRNA level (Figure 1B) suggests that the IRF1 promoter in HIV-R is “primed” for silencing in response to a stimulus. This is certainly not the first study to show HDAC association with an active gene locus.47 The activity of HDAC requires that it be assembled into larger protein complexes. How HDAC2 is recruited or regulated at IRF1 promoter is not clear and requires further study. HDAC2 has been reported to associate with NF-κB/p65 via dimerization with HDAC1,48 and its activity can be inhibited by acetylated HDAC1 protein.49 However, our ChIP experiment failed to detect HDAC1 binding to IRF1 promoter (data not shown). Nevertheless, these observations strongly support the notion that the IRF1 promoter in HIV-R individuals is “primed” via HDAC2 binding for a rapid silencing of IRF1 responses.
Heritable silencing of gene expression has been associated with the spreading of epigenetic modifications across gene locus.14 Here, we observed the spreading of HDAC2 binding and histone deacetylation across the IRF1 gene locus in HIV-R individuals (Figures 5A, 7B; supplemental Figure 2). It suggests that the epigenetic control of IRF1 response in HIV-R individuals is intrinsic, in that, if the IRF1 response were to be elicited again in vivo or in vitro, a similar response would be observed. This is the first study to show the involvement of HDAC2 in regulating IRF1 response to IFN-γ stimulation in primary PBMCs (Figure 7B) and the first to show that there were significant amounts of HDAC2 binding to the IRF1 promoter of ex vivo, unstimulated PBMCs (Figure 7A). Our ongoing studies will examine how reduced basal HDAC2 expression may affect IRF1 response induced by viral infection and in the clinical resistance against HIV-1 infection.
Resistance to HIV infection is a multifactorial phenomenon. Understanding what is protecting these women from HIV seems essential to developing effective HIV preventions, such as a vaccine or microbicide or other therapeutics. Our data provide convincing evidence that robust, but transient, IRF1 responses may be one of critical factors in restricting early HIV replication.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank J. Rutherford, S. Ramdahin, and L. Slaney for technical support, Dr K. Fowke for reviewing the manuscript, and the participants in the Pumwani cohort.
This work was supported by the National Institutes of Health (grant R01 AI56980 A1), the Canadian Institutes of Health Research (HOP-43135), the Bill and Melinda Gates Foundation, and the Canadian Institutes of Health Research through the Grand Challenges in Global Health Initiative (F.A.P.). F.A.P. is a Tier 1 Canada Research Chair in Susceptibility and Resistance to Infection.
National Institutes of Health
Authorship
Contribution: R.-C.S. and T.B.B. designed the research project and wrote the manuscript; R.-C.S. performed the majority of the experiments and analyzed the data; A.S. performed some experiments; J.K. and W.J. provided clinical and technical advice as well as subject characterization and recruitment; and F.A.P. provided funds to support the project.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ruey-Chyi Su, Department of Medical Microbiology and Infectious Diseases, University of Manitoba, 507-730 William Ave, Winnipeg, MB, Canada, R3E 0W3; e-mail: su@cc.umanitoba.ca.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal