Abstract
The identity of T-cell progenitors that seed the thymus has remained controversial, largely because many studies differ over whether these progenitors retain myeloid potential. Contradictory reports diverge in their use of various in vitro and in vivo assays. To consolidate these discordant findings, we compared the myeloid potential of 2 putative thymus seeding populations, common lymphoid progenitors (CLPs) and multipotent progenitors (MPPs), and the earliest intrathymic progenitor (DN1), using 2 in vitro assays and in vivo readouts. These assays gave contradictory results: CLP and DN1 displayed surprisingly robust myeloid potential on OP9-DL1 in vitro stromal cocultures but displayed little myeloid potential in vivo, as well as in methylcellulose cultures. MPP, on the other hand, displayed robust myeloid potential in all settings. We conclude that stromal cocultures reveal cryptic, but nonphysiologic, myeloid potentials of lymphoid progenitors, providing an explanation for contradictory findings in the field and underscoring the importance of using in vivo assays for the determination of physiologic lineage potentials.
Introduction
The generation of mature hematopoietic lineages from stem cells entails the progressive loss of self-renewal capacity along with an increase in lineage restriction. Lineage restriction requires up-regulation of lineage-specifying genes and down-regulation of genes that promote alternate fates,1 probably involving progressive modulation of both transcription factor activity and chromatin structure. Accordingly, reinitiation of alternate transcriptional profiles should result in restoration of suppressed lineage potentials in cells that are not yet fully lineage-committed. Consistent with this model, signaling through ectopically expressed human interleukin-2Rα (IL-2Rα) or granulocyte-macrophage colony-stimulating factor receptor (GM-CSFR) in common lymphoid progenitors (CLPs) allows them to mature down a myeloid fate at the expense of lymphoid development.2,3 The observation that signaling through the same ectopic cytokine receptor is unable to reprogram fully T cell–committed thymic progenitors to a myeloid fate indicates that as lineage commitment occurs, fate decisions are indeed progressively locked in4 : however, even these commitment decisions are not irreversible. Ectopic expression of PU.1 or C/EBPα, which compete with Pax5 to determine myeloid versus lymphoid fates in CLP, restores myeloid potential in T-cell “committed” progenitors.5,6 Mature B cells can also be reprogrammed to become T cells by conditional deletion of Pax5.7 Strikingly, ectopic expression of 4 transcription factors reprograms differentiated fibroblasts to a pluripotent state.8,9 Thus, lineage potentials revealed by experimental modifications may not reflect physiologic lineage decisions.
Strikingly, culturing cells outside of their in vivo environments can also promote nonphysiologic lineage outcomes. For example, VCAM-1− multipotent progenitors (MPPs) are highly lymphoid biased in vivo but efficiently adopt a myeloid fate in vitro.10 Similar myeloid fates could be induced in vivo if the VCAM-1− MPPs were redistributed to different microenvironments, suggesting that factors sequestered at local niches impact lineage decisions. Because such factors may be provided ectopically in vitro, these assays may reveal lineage potentials not realized in vivo.
Recent studies, which relied heavily on in vitro assays, suggested that the majority of murine thymic DN1 cells retain myeloid and T, but not B, lineage potential.11,12 These studies contradicted previous work indicating that thymus seeding progenitors are lymphoid committed.13-15 To investigate whether assay differences accounted for these contradictory results, we compared lineage potentials of DN1, CLP, and MPP in vitro, as well as in vivo, and asked whether the myeloid potential of DN1 was physiologic or was a cryptic potential revealed by the particular in vitro assay used. We confirm that DN1 can efficiently generate myeloid cells in vitro on OP9:OP9-DL1 stromal cocultures; strikingly, CLPs have comparably robust myeloid potential in this system. In stark contrast, CLP and DN1 have minimal residual myeloid potential in multiple in vivo contexts. As expected, MPP are bipotent for lymphoid and myeloid outcomes in all settings assayed. Thus, the in vitro myeloid potential of DN1 results from ectopic culture-specific conditions and does not reflect in vivo precursor-progeny relationships between DN1 and upstream hematopoietic progenitors.
Methods
Mice
C57BL6/Ka and CD45 congenic strains were housed at Stanford University or at the Joslin Diabetes Center animal facilities. All experiments involving mice were approved by the Stanford and Joslin Institutional Animal Care and Use Committees. GFP-transgenic mice carry a transgene encoding enhanced GFP under the control of the actin promoter and have been backcrossed multiple generations onto the C57BL/6 background.16
Antibodies
Antibodies were either affinity purified from hybridoma supernatants and used directly or conjugated to fluorophores in house, or were purchased as direct conjugates from eBioscience or BD Biosciences PharMingen. Flow cytometry and fluorescence-activated cell sorting (FACS) were performed on a FACSAria (BD Biosciences) and analyzed with FlowJo software Version 8.8.6 (TreeStar).
Cell purification, sorting, and transplantation
Bone marrow suspensions were centrifuged through Histopaque 1119 (Sigma-Aldrich) to clear red cells and bone fragments. Bone marrow and thymocytes were stained with purified antibodies to lineage markers, depleted using antirat IgG-conjugated Dynal beads (Dynal), stained with Tricolor-conjugated antirat IgG (Caltag), blocked with free rat-IgG, and stained with fluorescently conjugated antibodies. The cell populations were defined phenotypically and sorted as: DN1-linlo CD4−CD8−CD44+CD25−c-Kit+; CLP-linloFlk2+CD27+IL-7R+; MPP-linloFlk2+CD27+IL-7R−c-Kit+Sca-1+. For in vivo analysis, 5000 sorted cells were injected retro-orbitally into sublethally irradiated (450 cGy with an x-ray irradiator) or lethally irradiated (2 split doses of 450 cGy) CD45 congenic mice, as indicated. Spleen, bone marrow, and thymus were analyzed at the indicated times after cell transfer.
OP9:OP9-DL1 cocultures
Double-sorted MPP, CLP, and DN1 were clonally sorted onto confluent wells of 1:1 OP9:OP9-DL1 stromal cells in minimal essential medium-α with 10% fetal bovine serum and 5 ng/mL Fms-like tyrosine kinase 3 ligand (Flt3L) and IL-7, and 10 ng/mL each stem cell factor (SCF), IL-3, IL-6, M-CSF, GM-CSF, and granulocyte colony-stimulating factor (G-CSF; PeproTech). After 11 days, donor-derived cells were characterized by flow cytometry.
Methylcellulose culture
Double-sorted MPP, CLP, and DN1 were plated on 1 mL Methocult M3231 (Stem Cell Technologies) supplemented with 5 ng/mL Flt3L, and 10 ng/mL each SCF, IL-3, IL-6, M-CSF, GM-CSF, and G-CSF (PeproTech) in Iscove modified Dulbecco medium. Colonies were counted at day 6.
Cytospins
After 11 days on OP9:OP9-DL1 cocultures, hematopoietic progeny from wells of 100 plated MPP, CLP, and DN1 cells were analyzed by flow cytometry, sorted, cytospun, and stained with Hemacolor (EMD Chemicals). Images were obtained with a Nikon Eclipse E800 microscope, using a SPOT RTSlider model 2.3.1 camera with Diagnostic Instrument SPOT software Version 4.5.9.6, with a 20×/.75 NA objective.
Results
DN1 and CLP display robust myeloid potential on OP9:OP9DL1 cocultures, but not in vivo
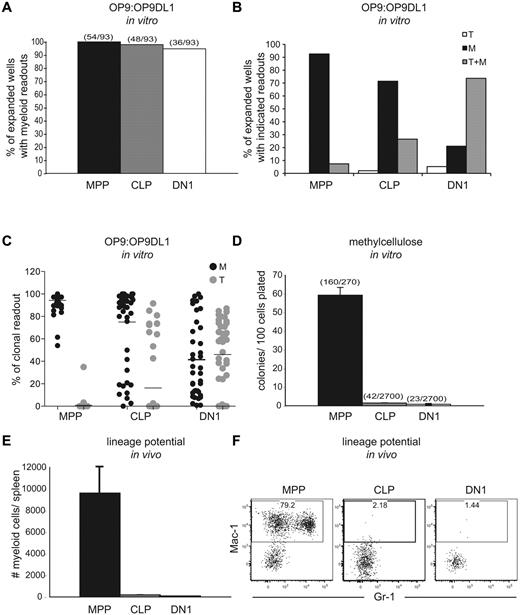
To initially compare lymphoid and myeloid potentials of putative T-cell progenitor populations, we cultured single MPP, CLP, or DN1 cells on a 1:1 mixture of OP9 and OP9DL1 cell lines in the presence of myeloid and lymphoid-promoting cytokines. In agreement with previous reports, after 11 days, 95% of wells expanded from DN1s had myeloid progeny (Figure 1A), and MPP yielded high clonal myeloid readouts, as expected.11,12 Notably, 98% of the expanded wells seeded with CLP also contained myeloid cells. Such a pronounced myeloid potential from CLP was unexpected because lymphoid restriction of this population in vivo had been observed in several other studies.13,15,17,18
The substantial in vitro myeloid potential of DN1 cells is not reproduced in either methylcellulose assays or in vivo. (A) MPP, CLP, or DN1 cells were clone- sorted into 96-well plates containing a confluent layer of a 1:1 mixture of OP9:OP9DL1 stroma in the presence of Flt3L, SCF, IL-3, IL-6, M-CSF, GM-CSF, G-CSF, and IL-7. After 11 days, wells were analyzed by flow cytometry for lineage markers, and those with cells of the phenotype CD45.2+ CD25− Mac-1+ were scored as myeloid cells. Graphs show the percentage of expanded colonies that contained at least 1% myeloid cells within the hematopoietic readout. Parentheses above columns indicate the number of wells with myeloid readout out of the total number of wells plated (regardless of whether the sorted cells expanded). The clonal readouts were composed almost entirely of either T lineage cells or myeloid lineages. Dendritic cells, as defined by CD11c+ Gr1−, made up a minor component of the myeloid readout (Figure 2). Results are representative of 2 independent experiments. (B) Expanded wells from the assay in panel A were scored for colonies that contained T cells only, myeloid cells only, or T cells and myeloid cells. T cells were defined as CD45.2+ CD25+, and myeloid cells were defined as in panel A. The colonies had to contain at least 1% myeloid or T readouts to be scored for the respective lineage. (C) The percentage of myeloid or T cells, as defined earlier in the Figure 1 legend, in all wells with clonal expansion from the assay in panel A. (D) A total of 135 MPP, 1350 CLP, and 1350 DN1 cells were FACS-sorted into methylcellulose containing Flt3L, SCF, IL-3, IL-6, M-CSF, GM-CSF, and G-CSF in 2 separate experiments. After 6 days, colonies were counted, and the number of colonies per 100 cells plated was scored. Parentheses above columns indicate the total number of colonies per number of cells plated in the summation of the 2 experiments. Error bars represent the SD between plates. (E) A total of 5000 MPP, CLP, or DN1 cells were sorted from CD45.1 congenic donors and injected intravenously into sublethally irradiated recipients. After 7 days, splenic chimerism and lineages were assessed by flow cytometry. The graph shows the absolute number of donor-derived myeloid cells (CD45.1+Mac-1+CD11c−) per spleen. Error bars represent the SD between mice. (F) Representative flow cytometric plots of donor splenocytes, showing myeloid readout quantified in panel E. Plots are pregated for CD45.1+ (donor) and CD11c− cells.
The substantial in vitro myeloid potential of DN1 cells is not reproduced in either methylcellulose assays or in vivo. (A) MPP, CLP, or DN1 cells were clone- sorted into 96-well plates containing a confluent layer of a 1:1 mixture of OP9:OP9DL1 stroma in the presence of Flt3L, SCF, IL-3, IL-6, M-CSF, GM-CSF, G-CSF, and IL-7. After 11 days, wells were analyzed by flow cytometry for lineage markers, and those with cells of the phenotype CD45.2+ CD25− Mac-1+ were scored as myeloid cells. Graphs show the percentage of expanded colonies that contained at least 1% myeloid cells within the hematopoietic readout. Parentheses above columns indicate the number of wells with myeloid readout out of the total number of wells plated (regardless of whether the sorted cells expanded). The clonal readouts were composed almost entirely of either T lineage cells or myeloid lineages. Dendritic cells, as defined by CD11c+ Gr1−, made up a minor component of the myeloid readout (Figure 2). Results are representative of 2 independent experiments. (B) Expanded wells from the assay in panel A were scored for colonies that contained T cells only, myeloid cells only, or T cells and myeloid cells. T cells were defined as CD45.2+ CD25+, and myeloid cells were defined as in panel A. The colonies had to contain at least 1% myeloid or T readouts to be scored for the respective lineage. (C) The percentage of myeloid or T cells, as defined earlier in the Figure 1 legend, in all wells with clonal expansion from the assay in panel A. (D) A total of 135 MPP, 1350 CLP, and 1350 DN1 cells were FACS-sorted into methylcellulose containing Flt3L, SCF, IL-3, IL-6, M-CSF, GM-CSF, and G-CSF in 2 separate experiments. After 6 days, colonies were counted, and the number of colonies per 100 cells plated was scored. Parentheses above columns indicate the total number of colonies per number of cells plated in the summation of the 2 experiments. Error bars represent the SD between plates. (E) A total of 5000 MPP, CLP, or DN1 cells were sorted from CD45.1 congenic donors and injected intravenously into sublethally irradiated recipients. After 7 days, splenic chimerism and lineages were assessed by flow cytometry. The graph shows the absolute number of donor-derived myeloid cells (CD45.1+Mac-1+CD11c−) per spleen. Error bars represent the SD between mice. (F) Representative flow cytometric plots of donor splenocytes, showing myeloid readout quantified in panel E. Plots are pregated for CD45.1+ (donor) and CD11c− cells.
The OP9:OP9DL1 cultures promoted T-cell differentiation as well, although less efficiently than they promoted myeloid differentiation. Less than 10% of MPP-seeded wells yielded T lineage cells, whereas CLPs and DN1 differentiated into T cells with increasing efficiency (Figure 1B-C). Nevertheless, the efficiency of this culture system in driving myeloid differentiation was illustrated by the fact that, regardless of the progenitor used, less than 10% of the wells yielded only T cells, and almost all wells that contained T cells also contained myeloid cells. Furthermore, the average percentage of myeloid cells in clonally expanded wells seeded with DN1 was approximately 41%, ranging up to 100% (Figure 1C). Therefore, the OP9:OP9DL1 system robustly promotes myeloid differentiation.
We next assessed the myeloid potential of CLP, DN1, and MPP on methylcellulose in the presence of myeloid-promoting cytokines.19 Interestingly, MPP efficiently generated myeloid colonies (59%), whereas neither CLP nor DN1 had substantial colony-forming ability (both < 2%; Figure 1D). Because both methylcellulose and OP9:OP9DL1 cultures contained the same concentrations of myeloid-promoting cytokines, the discrepant readouts suggest that other factors drive myeloid development in the OP9:OP9DL1 culture.
We also examined the in vivo lineage potentials of MPP, CLP, and DN1 by analyzing multilineage splenic chimerism 7 days after transplantation into sublethally irradiated recipients. MPP yielded substantial myeloid progeny, with 79% of donor-derived CD11c− cells expressing the myeloid marker Mac-1. However, neither CLP nor DN1 generated more than rare myeloid progeny in vivo (Figure 1E-F), starkly contrasting with their robust myeloid potential in the stromal coculture system.
Multiple types of myeloid progeny are produced on OP9:OP9DL1 cocultures
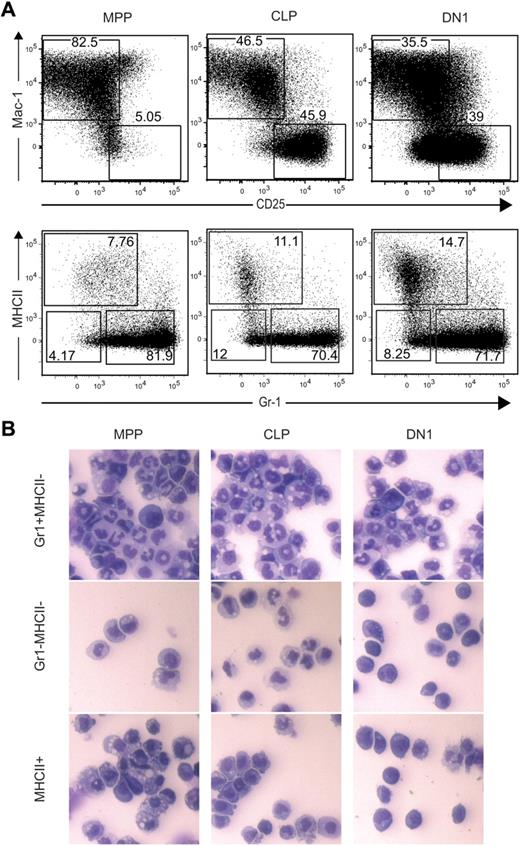
We further characterized myeloid progeny of MPP, CLP, and DN1 on OP9:OP9DL1 cultures to determine whether they differentially produced specific myeloid subtypes. Mac-1+ progeny could be subdivided into Gr-1+MHCII−, Gr-1−MHCII−, and MHCII+ populations (Figure 2A). The MHCII+ cells were largely CD11c+CD80+ dendritic cells. The MHCII−Gr-1− and the MHCII−Gr-1+ cells had characteristics of macrophages and granulocytes, respectively. By cytospin, the MHCII−Gr-1+ cells displayed the multilobed nuclei of granulocytes, whereas the MHCII−Gr-1− cells had the large vacuolated cytoplasm of macrophages (Figure 2B). The MHCII+ population contained some cells with cytoplasmic projections consistent with dendritic cells, although MPP-derived cells had vacuoles consistent with phagocytic activity. In summary, MPP, CLP, and DN1 were all able to generate similarly diverse myeloid subtypes on OP9:OP9-DL1 cocultures, although granulocytes predominated over dendritic cells and macrophages.
The OP9:OP9DL1 culture system supports development of multiple myeloid lineages from MPP, CLP, and DN1. (A) A total of 100 MPP, CLP, or DN1 cells were sorted into 24-well plates containing a confluent layer of a 1:1 mixture of OP9:OP9DL1 stroma in the presence of Flt3L, SCF, IL-3, IL-6, M-CSF, GM-CSF, G-CSF, and IL-7. After 11 days, wells were analyzed by flow cytometry. Upper plots are gated on donor-derived cells and show gates for Mac-1+CD25− myeloid cells and CD25+Mac-1− T cells; lower plots are gated on the donor-derived, Mac-1+CD25− cells from the upper plots, revealing 3 subpopulations of myeloid cells generated from all 3 sorted progenitors. (B) The Gr-1+MHCII−, Gr-1−MHCII−, and MHCII+ myeloid subsets shown in panel A were sorted, plated onto slides using a cytospin, and analyzed morphologically by Hemacolor stains.
The OP9:OP9DL1 culture system supports development of multiple myeloid lineages from MPP, CLP, and DN1. (A) A total of 100 MPP, CLP, or DN1 cells were sorted into 24-well plates containing a confluent layer of a 1:1 mixture of OP9:OP9DL1 stroma in the presence of Flt3L, SCF, IL-3, IL-6, M-CSF, GM-CSF, G-CSF, and IL-7. After 11 days, wells were analyzed by flow cytometry. Upper plots are gated on donor-derived cells and show gates for Mac-1+CD25− myeloid cells and CD25+Mac-1− T cells; lower plots are gated on the donor-derived, Mac-1+CD25− cells from the upper plots, revealing 3 subpopulations of myeloid cells generated from all 3 sorted progenitors. (B) The Gr-1+MHCII−, Gr-1−MHCII−, and MHCII+ myeloid subsets shown in panel A were sorted, plated onto slides using a cytospin, and analyzed morphologically by Hemacolor stains.
Only MPP differentiate efficiently into myeloid lineages in vivo
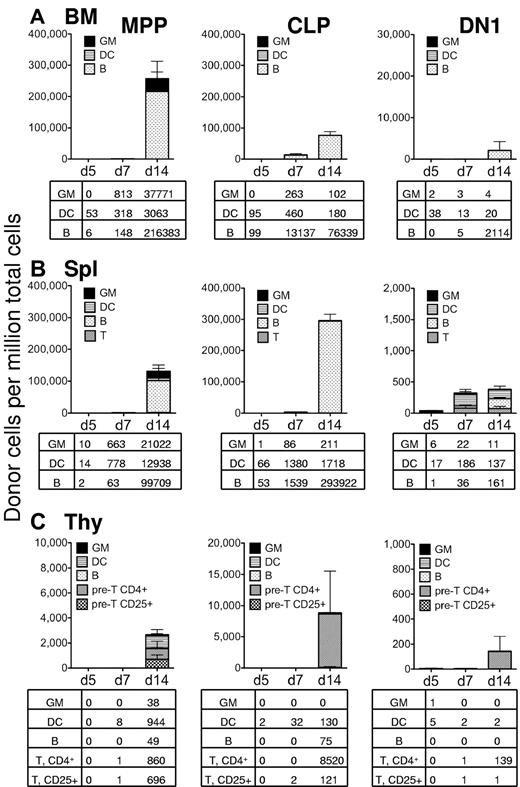
Our initial in vivo analysis measured progenitor differentiation into myeloid lineages only within the spleen, 7 days after transplantation, leaving open the possibility that CLP and DN1 could generate myeloid progeny in vivo in different tissues or at a different time points. Therefore, we investigated lineage output from 5000 intravenously injected MPP, CLP, and DN1 over 5 to 14 days in the bone marrow, spleen, and thymus. Fourteen days after transplantation into sublethally irradiated recipient mice, MPP generated approximately 40 000 Mac-1+Gr-1+ granulocytes per million bone marrow cells (Figure 3A). Neither CLP nor DN1 generated substantial granulocyte chimerism within the marrow cavity (Figure 3A). Instead, CLP differentiated primarily into CD19+ B cells in the bone marrow by days 7 to 14, indicating both their lymphoid restriction and their ability to seed the bone marrow. DN1, while yielding very low bone marrow chimerism at days 5 and 7, did differentiate into some B cells by day 14, showing that the population of DN1 cells maintains some B-cell potential, as had been previously described.20 MPP also produced B cells in the bone marrow, as expected for a lymphoid/myeloid bipotent progenitor. MPP, CLP, and DN1 all generated low numbers of CD11c+ dendritic cells in the bone marrow (Figure 3A).
In sublethally irradiated recipients, only MPPs display robust myeloid potential. Sublethally irradiated recipient C57BL/6 mice were transplanted with 5000 MPP (left column), 5000 CLP (middle column), or 5000 DN1 cells (right column) from GFP-transgenic mice. After 5, 7, or 14 days, the bone marrow (A), spleen (B), and thymus (C) of each mouse was analyzed for GFP+ donor cells by flow cytometry. Each column represents the level of GFP+ chimerism in that tissue, and the different patterns indicate the chimerism of the different lineages. The lineages were determined as follows: Myeloid cells (GM) were defined as Mac-1+Gr-1+CD11c−; dendritic cells (DC) were defined as CD11c+; B cells were defined as CD19+. T lineage cells were defined as either CD4+ (pre-T, CD4+) or CD25+ (pre-T, CD25+) in the thymus. At day 14, most thymocytes were immature, and antibodies to CD4 and CD25 labeled nearly all T lineage cells. The tables below each graph show the frequency (cells/million live cells) of each donor cell lineage for each tissue analyzed and represent the average from 3 transplanted mice. This experiment was performed twice, with similar results.
In sublethally irradiated recipients, only MPPs display robust myeloid potential. Sublethally irradiated recipient C57BL/6 mice were transplanted with 5000 MPP (left column), 5000 CLP (middle column), or 5000 DN1 cells (right column) from GFP-transgenic mice. After 5, 7, or 14 days, the bone marrow (A), spleen (B), and thymus (C) of each mouse was analyzed for GFP+ donor cells by flow cytometry. Each column represents the level of GFP+ chimerism in that tissue, and the different patterns indicate the chimerism of the different lineages. The lineages were determined as follows: Myeloid cells (GM) were defined as Mac-1+Gr-1+CD11c−; dendritic cells (DC) were defined as CD11c+; B cells were defined as CD19+. T lineage cells were defined as either CD4+ (pre-T, CD4+) or CD25+ (pre-T, CD25+) in the thymus. At day 14, most thymocytes were immature, and antibodies to CD4 and CD25 labeled nearly all T lineage cells. The tables below each graph show the frequency (cells/million live cells) of each donor cell lineage for each tissue analyzed and represent the average from 3 transplanted mice. This experiment was performed twice, with similar results.
In the spleens of sublethally irradiated recipients, MPP generated Mac-1+Gr-1+ granulocytes from days 7 to 14, in addition to producing B cells and dendritic cells by day 14 (Figure 3B). Whereas MPP produced approximately 20 000 granulocytes per million splenocytes at day 14, CLP generated only approximately 200 Mac-1+Gr-1+ cells, and DN1 generated only 10 granulocytes at this time, again indicating the low myeloid potential of these cell types in vivo. CLP made primarily B cells, demonstrating their potential to produce abundant numbers of lymphocytes, and DN1, although the chimerism was low, yielded primarily B cells and dendritic cells.
Thymic chimerism was low from all 3 populations 5 to 14 days after intravenous injection into sublethally irradiated recipients, and only MPP gave rise to any myeloid progeny within the thymus. Even myeloid chimerism from MPP was proportionally low in the thymus and significantly lower than dendritic cell, B cell, or T lineages (Figure 3C). CLP did not give rise to thymic myeloid cells, although they did yield approximately 8500 CD4+ thymocytes per million cells. The finding that CLP yielded higher chimerism at day 14 from CLP relative to MPP is consistent with our previous studies, showing that MPP give rise to CLP, which are then the major bone marrow proximate source of T-cell progenitors.15 DN1 gave extremely low thymic readouts at all time points, indicating that they lose the potential to reseed the thymus efficiently.
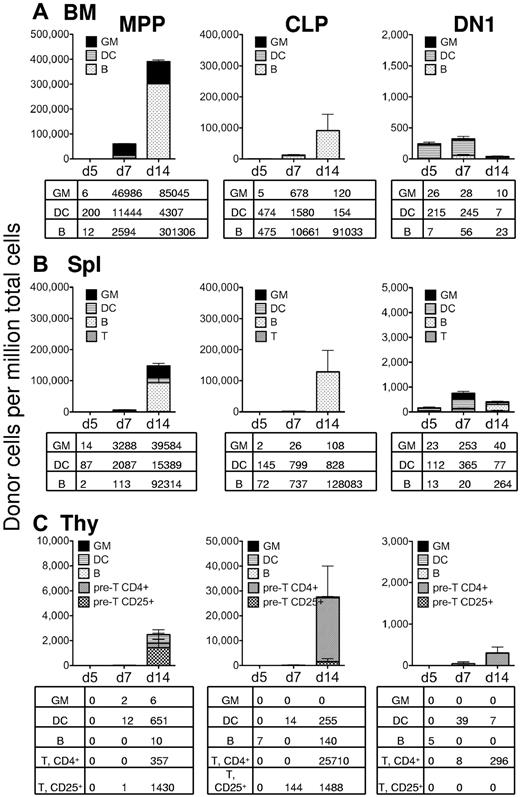
Although MPP revealed much higher myeloid chimerism in the spleen and bone marrow of sublethally irradiated recipients compared with CLP and DN1, it was possible that a lethally irradiated recipient would have an altered microenvironment, rendering it more conducive to myelopoiesis from these progenitors. Therefore, we also analyzed lineage potentials in the spleen, bone marrow, and thymus of lethally irradiated recipients. In the bone marrow, MPPs were still by far the major producers of granulocytes both at day 7 and day 14, and CLP yielded a large B-cell readout at these time points (Figure 4A). Dendritic cell chimerism appeared to be slightly faster and higher in lethally versus sublethally irradiated recipients from all 3 cell types (Figures 3A, 4A). Similarly, in the spleen, MPP generated approximately 40 000 granulocytes per million splenocytes in 14 days compared with approximately 100 from CLP and approximately 40 from DN1 per million cells (Figure 4B). However, CLP generated significant numbers of B cells by day 14, and DN1 generated some splenic dendritic cells at day 7, indicating that both populations could yield splenic readouts. Finally, in the thymus of lethally irradiated recipients, none of the 3 cell types yielded substantial myeloid chimerism, although all 3 generated T lineage readout and dendritic cells (Figure 4C). In summary, these data support lymphoid commitment of CLP and DN1 cells in vivo, whereas MPPs alone have robust myeloid potential.
In lethally irradiated recipients, only MPP display robust myeloid potential. Lethally irradiated recipient mice were transplanted with 5000 MPP (left column), 5000 CLP (middle column), or 5000 DN1 (right column) from GFP-transgenic mice. Recipient mice were also transplanted with 300 000 “helper” bone marrow cells, so that they would recover from the irradiation. Other than the irradiation dosage and the use of “helper” bone marrow, the description of this figure is identical to the description of Figure 3. (A) Bone marrow cells. (B) Splenocytes. (C) Thymocytes.
In lethally irradiated recipients, only MPP display robust myeloid potential. Lethally irradiated recipient mice were transplanted with 5000 MPP (left column), 5000 CLP (middle column), or 5000 DN1 (right column) from GFP-transgenic mice. Recipient mice were also transplanted with 300 000 “helper” bone marrow cells, so that they would recover from the irradiation. Other than the irradiation dosage and the use of “helper” bone marrow, the description of this figure is identical to the description of Figure 3. (A) Bone marrow cells. (B) Splenocytes. (C) Thymocytes.
Discussion
In this study, we find that factors present in the OP9:OP9DL1 culture system can promote myelopoiesis of DN1 and CLP; however, this high-level in vitro myeloid potential is not recapitulated in multiple in vivo settings. The contrast between the myeloid potentials revealed with in vitro stromal cocultures and in vivo transplantation assays strongly suggests that experiments designed to determine hematopoietic lineage fate maps should use assays that most closely represent the in vivo situation. Although transplantation itself could create artifacts, a recent lineage-tracing experiment, in which progeny of cells expressing IL-7 receptor were irreversibly marked with a fluorescent reporter, indicated that in untransplanted mice, CLP-derived lineages were almost entirely restricted to lymphoid fates.14 The agreement between lineage tracing and transplantation of CLP, reported here and elsewhere, supports transplantation as a strong tool for determining in vivo lineage hierarchies.
The stromal coculture assays, although not reflective of in vivo fates, do indicate that CLP and DN1 have the ability to give rise to myeloid cells under certain conditions. Therefore, the mechanism of lineage fate restriction in vivo is not sufficient to maintain lymphoid lineage dominant commitment in vitro. However, it is possible that under certain circumstances the in vitro myeloid potential of CLP and DN1 could be revealed in vivo. We did find, for example, that lethal irradiation caused an increase in the number of myeloid cells derived from both CLP and DN1. However, the number of myeloid progeny was still very small, in the range of 1000 times less than from MPP under the same conditions. It is possible that other stressful conditions, such as infection, could also alter the lineage outcomes of CLP and DN1, but under normal physiologic conditions, these cell types do not yield substantial numbers of myeloid progeny.
Several caveats should be considered when interpreting the lack of substantial in vivo myeloid readout from CLP and DN1. First, low myeloid chimerism might have simply reflected the failure of CLP or DN1 to engraft productively in the host. We found high and consistent levels of donor chimerism in animals transplanted with MPP and CLP. CLP generated large numbers of B cells in the spleen and bone marrow of recipients but did not yield a large myeloid readout. Thus, the lymphoid restriction of CLP on transplantation is not a reflection of poor chimerism. DN1, in contrast, engrafted relatively poorly. One probable cause of poor DN1 engraftment is that CLP-derived DN1 cells lose thymic-homing potential but require thymus-derived growth signals to survive in vivo. Nevertheless, the relative myeloid chimerism among engrafted DN1 cells was not high at any time point examined: instead, lymphoid and/or dendritic chimerism was always higher. Thus, neither DN1 nor CLP shares the high level myeloid potential of MPP in vivo.
A second caveat that could account for lack of myeloid progeny in vivo is that CLP and DN1 may lack homing receptors to recruit them to a myeloid-inducing environment. Arguing against this possibility is the observation that CLP and DN1 also do not efficiently take on myeloid fates in methylcellulose cultures, where they are bathed with myeloid-inducing factors. Furthermore, because DN1 and CLP were able to generate myeloid progeny efficiently in stromal cocultures, their inability to yield substantial colonies in methylcellulose assays did not reflect poor viability after sorting. Rather, their failure to generate myeloid methylcellulose colonies suggests that, instead of homing issues, CLP and DN1 probably have lost their ability to efficiently respond to myeloid-driving cytokines and factors, either through down-regulation of cytokine receptors, intracellular signaling components, or transcriptional networks.
The cryptic myeloid potential of CLP revealed by OP9:OP9DL1 cocultures was recently confirmed in a study that also solidified the IL-7R+ origin of the majority of DN1s.14 Because IL-7R is first expressed on CLP in the bone marrow, this finding is consistent with our previous report of a CLP-DN1 precursor-progeny relationship.15 Consistent with both of these studies, we find here that, compared with MPP, CLP yield B cells and T lineage cells with faster kinetics in the bone marrow and thymus, respectively (Figures 3, 4). If DN1 were derived directly from MPP, which retain myeloid potential in vivo, one would expect faster T lineage chimerism from MPP relative to CLP, whereas we see faster T lineage chimerism from CLP.
In conclusion, DN1 and CLP have initiated lymphoid lineage commitment but have not irreversibly silenced myeloid potential, as demonstrated in OP9-OP9DL1 cocultures. This 2-step model of initial commitment to lymphopoiesis, revealed by in vivo transplantation, followed by final closure of myelopoietic potential, is essentially the same as we proposed previously.3 Lymphoid lineage specification entails progressive changes in gene transcription, influenced by epigenetic changes, such as histone modifications, and DNA methylation. In a recent study, we have found direct evidence that multiple genes, such as Meis1, Gcnt2, and Dach1, are progressively methylated and silenced as cells differentiate from uncommitted MPP through CLP, DN1, DN2, and T lineage-committed DN3 stages.21 Accordingly, myeloid gene loci may remain accessible during the initiation of lymphoid specification, Thus, one potential mechanism by which stromal cocultures may drive ectopic myelopoiesis would be to provide cellular and/or soluble factors able to reinitiate transcription from incompletely silenced myeloid-promoting loci. If these factors were not normally present in vivo, perhaps because of microenvironmental restrictions, comparable myeloid fates would not be realized under physiologic conditions. Our results highlight the discrepancies in lineage readouts between in vivo and in vitro assays and emphasize the importance of using in vivo assays to characterize/confirm physiologic lineage hierarchies.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Matthew Inlay for helpful comments, Christina Muscat and Teja Naik for antibodies, Adriane Mosley for help with mice, Libuse Jerabek for facilitating experiments, Erika Brooks for excellent technical assistance, and J. LaVecchio and G. Buruzula at Joslin Diabetes Endocrinology Research Center/Harvard Stem Cell Institute Flow Cytometry Core.
This work was supported by the Leukemia & Lymphoma Society (Special Fellow Career Development award; L.I.R.E.) and the National Institutes of Health grants 5R01A1047457 and 5R01A1047458 (to I.L.W.) and Joslin Diabetes Endocrinology Research Center grant P30DK36836 (to T.S.).
National Institutes of Health
Authorship
Contribution: L.I.R.E. and T.S. designed, implemented, and analyzed the experiments jointly and prepared the manuscript jointly; and I.L.W. supervised the research project and contributed to the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation of L.I.R.E. is Institute of Cellular and Molecular Biology, Molecular Genetics and Microbiology, University of Texas at Austin, Austin, TX.
Correspondence: Lauren I. Richie Ehrlich, Molecular Genetics and Microbiology, 2506 Speedway, NMS 2.314, Austin, TX 78712; e-mail: lehrlich@mail.utexas.edu.
References
Author notes
L.I.R.E and T.S. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal