Abstract
An increasing body of data has demonstrated that the traditional concept of morphologic complete remission in acute myeloid leukemia, in which less than 5% myeloblasts is regarded as a sufficient response criterion, is not biologically sound. Fortunately, the quantitative reverse-transcribed polymerase chain reaction (RT-PCR) method seems to be a promising alternative because of its high degree of preclinical standardization and extreme sensitivity on the background of an accurate day-to-day estimate of sample quality. Widespread implementation of this has, however, to some extent been hampered by the lack of knowledge of how and when to measure minimal residual disease levels and, even more importantly, how to react preemptively on a molecular relapse defined by a PCR reversal. Thus, only few prospective studies have been published to date to clinically validate this assay. Here, we discuss outstanding issues in the clinical implementation of RT-PCR for fusion transcripts, mutated and overexpressed genes in acute myeloid leukemia patients in complete remission, and propose a set of guidelines, which can be used when designing prospective trials aimed at validating the use of RT-PCR as well as for following these patients based on mathematical models for disease recurrence recently developed in our laboratory.
Introduction
Classification of acute myeloid leukemia (AML) is now increasingly based on biologically highly relevant genetic abnormalities, such as the recurrent genetic abnormalities, including balanced translocations t,(8,21), inv(16)/t,(16,16) t(15;17), t(6,9) inv(3)/t(3,3) t(1;22), other cytogenetic findings, or mutated genes (FLT3, NPM1 and CEBPA).1 The reason that such a classification has superseded morphologic and immunophenotypic ones like the French-American-British is that they entail clear implications for prognosis as evidenced in several seminal papers, summarized in Schlenk et al.2
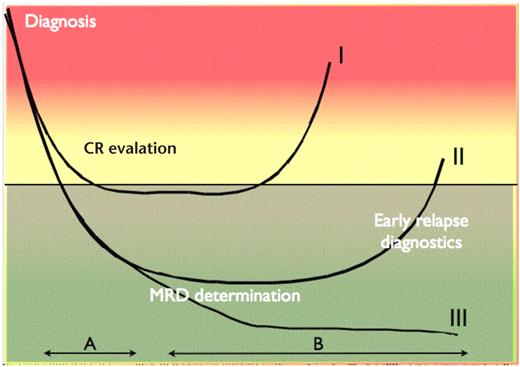
Ideally, these advances in characterizing AML patients should result in a platform for risk-adapted treatment for the majority of patients. Although such an approach has been hinted in recent World Health Organization classifications of the disease,1 it must be realized that these basic advances in our understanding of AML biology has only to a limited extent been accompanied by improved survival. This is because the majority of patients who obtain complete remission (CR) relapse from a state of “deep hematologic CR” depicted in Figure 1.3 In this context, the term minimal residual disease (MRD) was developed to denote the source of relapse in those patients, in whom no detectable disease was present by standard diagnostic tests, but who did nonetheless experience a recurrence of the leukemic clone.3
The MRD concept. x-axis represents time; y-axis represents tumor burden. The horizontal black line represents the sensitivity of standard morphologic analyses, such as light microscopy using immunohistochemical methods. I, II, and III indicate 3 different courses of disease documented by MRD measurements. Patient course I (early relapse) can be distinguished from II and III (late relapse and complete cytogenetic response, respectively) by MRD measurements during therapy (period A). Relapse (patient course I and II) can in some cases be identified several months before clinical symptoms by MRD measurements after discontinuation of therapy (period B).
The MRD concept. x-axis represents time; y-axis represents tumor burden. The horizontal black line represents the sensitivity of standard morphologic analyses, such as light microscopy using immunohistochemical methods. I, II, and III indicate 3 different courses of disease documented by MRD measurements. Patient course I (early relapse) can be distinguished from II and III (late relapse and complete cytogenetic response, respectively) by MRD measurements during therapy (period A). Relapse (patient course I and II) can in some cases be identified several months before clinical symptoms by MRD measurements after discontinuation of therapy (period B).
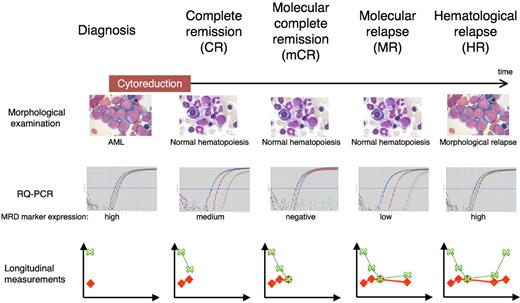
These observations in turn imply that the traditional hematologic relapse concept encompassing declining hematopoiesis and subsequent symptoms related to bone marrow failure need to be reevaluated and new terminologies defined. In Figure 2, we define these and will use these terms in the remainder of this review.
Terminology in MRD measurements. Definitions are based on the presence of blasts on light microscopy examination and disappearance of RT-PCR positivity in a sample with an acceptable sensitivity. RT-PCR panels represent raw data from MRD measurements of a patient using 2 control genes (red and blue curves) and one MRD marker (green curve). Longitudinal measurements represent MRD reporting as recommended by European Leukemia Network4 showing the MRD level (green crosses and lines) as well as the sample day-to-day sensitivity (red diamonds and lines).
Terminology in MRD measurements. Definitions are based on the presence of blasts on light microscopy examination and disappearance of RT-PCR positivity in a sample with an acceptable sensitivity. RT-PCR panels represent raw data from MRD measurements of a patient using 2 control genes (red and blue curves) and one MRD marker (green curve). Longitudinal measurements represent MRD reporting as recommended by European Leukemia Network4 showing the MRD level (green crosses and lines) as well as the sample day-to-day sensitivity (red diamonds and lines).
We think that there are now ample data in the cumulated literature to suggest a way of applying MRD testing in adult AML patients in molecular complete remission (mCR). This is, however, not the same as stating that its value for the patients and caretakers has been resolved, and the purpose of this Perspective is therefore: (1) to delineate the present state of MRD testing by quantitative reverse-transcribed polymerase chain reaction (RT-PCR) focusing on pros and cons; and (2) based on very recent data, to present a series of recommendations for sampling for the most commonly seen molecular aberrations in AML, which are amenable to this methodology. Applied collectively, these assays will result in a situation, in which more than 80% of adult AML patients can be offered MRD evaluation in a rational fashion. This should in turn ensure optimal results in future randomized trials, which will hopefully provide the ultimate evidence for the beneficial effect of individualized, molecularly based preemptive cytoreduction in AML patients.
Methods applicable for MRD detection
In theory, at least, any method with sensitivity higher than that of cytochemistry-assisted light microscopy can be used for MRD detection. However, methods available in the late 1980s and early 1990s (eg, semiquantitative competitive PCR) were only reliable when performed in highly specialized laboratories and, although groundbreaking in chronic myeloid leukemia (CML), never had much real impact on clinical practice in AML.
The development and validation of methods that allow for high degree of automation and sample throughput, such as fluorescence in situ hybridization, real-time, fluorescence-based quantitative RT-PCR, and multicolor flow cytometry, dramatically changed this. Each technique should be applied judicially and according to its advantages and drawbacks, and the reader is referred to recent literature regarding this. There is, however, no doubt that a validated flow cytometry assay with a sensitivity of 1:10 000 is an excellent tool for fast MRD evaluation in the daily clinical situation. Its obvious drawbacks include lack of specificity and of constancy over time of the aberrant antigen expression delineated at diagnosis. In contrast, the RT-PCR assay takes a longer time to perform and entails a considerably longer turnover time. On the other hand, there is no doubt that it is more specific and, for most targets, more sensitive than multiparameter flow cytometry. For this review, we will limit our deliberations to the RT-PCR methodology based on the fact that this is at the present by far the most preclinically validated assay among these.
Pros and cons of MRD detection by RT-PCR
RT-PCR combines the high sensitivity of the standard PCR assay with a fluorophore that emits light on amplification of the PCR product. The PCR cycle at which the PCR product-specific fluorescence can be distinguished from the background fluorescence is inversely proportional to the amount of the target sequence at the start of the reaction.5 This proportionality spans up to 6 log decades and thus imbues the method with a very high theoretical sensitivity. The target of the RT-PCR assay can be any DNA or RNA sequence present in the malignant cells. Thus, in cells where balanced translocations are present, fusion genes can be used as the MRD marker, either at the DNA or RNA level. Similarly, in cells with mutated genes (eg, mutated NPM1 [designated NPM1c]6 ), these can be used to detect the malignant cells.
A major advantage to the RT-PCR technique is the extensive preclinical optimization, which it has been afforded, either through single laboratory contributions, but to a high degree also by multicenter efforts. Thus, in a European Union–sponsored Europe Against Cancer initiative, several key features inherent to the assay were addressed, including the delineation of the control genes, which are crucial in determining the day-to-day quality of the assay.7 In addition, careful selections of probes and primers, which are now in widespread use, have resulted in robust reactions for several fusion transcripts seen in AML, acute lymphoblastic leukemia, and CML.4 Finally, within the auspices of the LeukemiaNet collaborative (http://www.leukemia-net.org), a reporting software package has been developed, which should enable harmonization of data presentation to clinical doctors and even patients, thus furthering the bedside clinical impact of RT-PCR results (Østergaard et al, Leukemia, in press).
However, it must be realized that several caveats remain before routine clinical implementation of the assay. First and foremost, the exceedingly high theoretical sensitivity seen in the diluted (“spiked”) cell line experiments reported in pioneering RT-PCR studies8-11 does not translate into similar ones when patient material is targeted. Thus, for many of the reactions, a sensitivity of not more than 1:10 000 on diagnosis samples is the routine rather than the exception (Table 1). This is partly because the expression levels of target transcript are rarely as high as in leukemic cells as in cell lines,4 partly because of the fact that some degradation of target transcript en route to the laboratory is inevitable, although this can be held at a minimum by adhering to standardized protocols for sample handling and storage.14 It is nevertheless important that all samples analyzed are interpreted in relation to the day-to-day sensitivity to show the end-user the real sensitivity instead of a theoretical one.
Quantitative RT-PCR MRD markers in AML
| Marker . | Chromosome(s) . | Frequency, % . | Sensitivity* . |
|---|---|---|---|
| Fusion transcripts | |||
| PML-RARA | t(15;17) | 712 | 1:5000-1:200 000 |
| RUNX1-RUNX1T1 | t(8;21) | 612 | 1:3000-1:500 000 |
| CBFB-MYH11 | inv(16) | 712 | 1:5000-1:100 000 |
| DEK-CAN | t(6;9) | 112 | 1:400-1:4000 |
| Overexpressed genes | |||
| WT1 | 11p13 | 8013 | 1:10-1:500/1:30-1:2000† |
| Mutated genes | |||
| NPM1c | 5q35 | 2513 | 1:10 000-1:100 000 |
| dupMLL | 11q23 | 613 | 1:100-1:2000‡ |
| FLT3-ITD | 13q12 | 2313 | 1:10 000 |
| Marker . | Chromosome(s) . | Frequency, % . | Sensitivity* . |
|---|---|---|---|
| Fusion transcripts | |||
| PML-RARA | t(15;17) | 712 | 1:5000-1:200 000 |
| RUNX1-RUNX1T1 | t(8;21) | 612 | 1:3000-1:500 000 |
| CBFB-MYH11 | inv(16) | 712 | 1:5000-1:100 000 |
| DEK-CAN | t(6;9) | 112 | 1:400-1:4000 |
| Overexpressed genes | |||
| WT1 | 11p13 | 8013 | 1:10-1:500/1:30-1:2000† |
| Mutated genes | |||
| NPM1c | 5q35 | 2513 | 1:10 000-1:100 000 |
| dupMLL | 11q23 | 613 | 1:100-1:2000‡ |
| FLT3-ITD | 13q12 | 2313 | 1:10 000 |
Sensitivity based on data in patient MRD follow-up performed at Laboratory of Immunohematology, Aarhus University Hospital and Munich Leukemia Laboratory (Susanne Schnittger, oral communication).
Sensitivity given as BM sensitivity/PB sensitivity and adjusted for expression in normal hematopoiesis.
Sensitivity adjusted for expression in normal hematopoiesis.
The most significant problem using RT-PCR is, however, the lack of internationally validated guidelines as to what clinical course of action to take when a molecular relapse (MR) is detected. This essential question will be dealt with in “A proposal for guidelines for the application of MRD assays in AML patients in CR.”
At least 2 more problems, both of biologic nature, need to be considered in relation to relapse evaluation. First, it should be kept in mind that development of therapy-related can mimic a relapse with loss of the original MRD markers and possibly acquisition of new ones. The exact frequency of such events is not well described (Jens Pedersen-Bjergaard, oral communication, November 2010), but might in some centers approach 5% (Susanne Schnittger, oral communication, November 2010). Second, a concern, which has been raised over the latest 2 decades for every new leukemia-associated disease marker developed, is the potential loss of marker expression between diagnosis and relapse resulting from clonal evolution. The problem is well described in the literature and known to be of concern both regarding RT-PCR-based15-21 and multicolor flow cytometry-based MRD follow-up.22,23 For RT-PCR, diligent construction of probes taking gene mutational hotspots as well as alternative splicing is taken into account aimed at avoiding the false-negative situations described in the literature,18,24,25 can reduce this problem to less than 5%.15,16,18,19 On the other hand, it is only fair to add that, as attractive as it looked initially, the FLT3 mutations have proven quite unstable and by and large unsuitable as a marker for MRD evaluation.
Target genes for determination of MRD in CR patients
The obvious choice for determining MRD in AML would be to assess for the genetic changes inherent to the malignant clone (eg, the fusion transcripts or mutated DNA sequences specific to the disease). Although some of these are quite frequent in AML (PML-RARA, CBFB-MYH11, RUNX1-RUNX1T1, NPM1 mutation type A and B), other aberrations are rare or are composed of several subtypes (TET2, FLT3-internal tandem duplications [ITD], and CEBPA). The latter necessitates the development of patient-specific primer-probe sets. Although this is possible, it is labor-consuming and makes standardization difficult. Thus, commonly known and extensively standardized MRD markers based on genetic changes account for no more than 35% of patients (Table 1) and overexpressed genes, such as Wilms tumor 1 (WT1) and PRAME, have been proposed and evaluated for use as MRD markers (Table 1). It is comforting in this respect that several studies have shown that these genes, despite their expression in healthy tissues, seem to be able to function as bona fide MRD targets despite their sensitivity being lower than for reactions targeting disease associated/specific genes.
Logistical and psychologic issues in performing MRD during CR
For those AML patients who unfortunately relapse, the amount of disease should increase gradually and be measurable by RT-PCR during the period preceding a hematologic relapse (HR; Figure 2). Supposedly, this MR will be first detectable in the bone marrow (BM), and only later in the peripheral blood (PB), although, paradoxically, this might not be true in all situations. Thus, irrespective of the caveats outlined herein, the high sensitivity of the RT-PCR methodology will deliver the opportunity to predict relapse weeks to months (in some cases, up to a year) before a clinical relapse occurs. This notion was indeed validated in several publications24-26 ; and given these observations taken together with the concomitant validation by the European Leukemia Network,6,7 it might seem strange that the method has not received more widespread use in individualized treatment of AML patients. The unresolved issue here is, most probably, that it is the practicing hematologist who will have to decide at the bedside whether to tell the patients about an RT-PCR conversion and, more importantly, to evaluate whether to act on it. Although we strongly believe in the concept of preemptive therapy based on RT-PCR data, we also realize that there is a barrier, which has to be crossed, when apparently healthy leukemia patients in CR are subjected to full-scale cytoreduction, entailing potentially lethal side effects. This, indeed, goes to the core of the Hippocratic Oath, which instructs us to first and foremost do no harm. It is therefore up to the researchers developing the MRD RT-PCR concept to provide the clinicians data supporting the use of preemptive therapy, for example, from randomized trials performed under the auspices of the Working Parties on Leukemia AML-17 protocol (Alan Burnett, e-mail communication, October 2009).
In addition to this psychologic barrier, it must be admitted that there are other outstanding issues that need to be addressed before implementation in clinical practice can be instituted in a fashion, which will convince the majority of clinicians. Thus: (1) Can overexpressed genes, encumbered with a lower sensitivity because of the expression in normal hematopoiesis, be used in post-therapy MRD measurements? (2) How long before HR can MRD really be detected for the different MRD markers presented in “Target genes for determination of MRD in CR patients,” and to what extent is this dependent on sampling intervals? In line with this: (3) How often should you sample patients to ensure a reasonable success in detecting MR? (4) To what extent can monitoring be accomplished by PB sampling alone, thus obviating the need for BM biopsies?
Definition of molecular relapse
Outside clinical trials, few clinicians will opt to institute treatments entailing high morbidity and mortality on the basis of MR determinations alone, fearing (as eluded to) the prospect of treating a cured patient exhibiting a temporary increase in a molecular marker of hitherto unknown significance. The question is therefore: how certain can one be that an HR will follow an MR and how can this notion be verified? First, there is the collected experience of the research community over the last decade following patients with no intervention and observing that this chain of events occurs in the majority of patients (ie, that MR is indeed followed by HR). Second, to look at this in a more formal fashion, we recently analyzed data on 500 AML patients in continuous CR and were able to establish cut-offs for when a positive MRD sample can be considered a MR for the 4 most common quantitative PCR MRD targets, namely, PML-RARA and CBFB-MYH11 (false-positive samplings rare, all positive measurement suspect of relapse), RUNX1-RUNX1T1 (cut-off level 10−4), and NPM1c+ (cut-off level 5 × 10−5).27 For overexpressed genes, such as WT1, the cut-off level has often been defined based on expression in normal hematopoiesis.16,28 Support for these data can be found in large studies for PML-RARA.29,30 Taken together, we think that these data allow for algorithms to be developed for aberration-specific sampling of AML patients in mCR in multicenter studies. Outside such, we think that the strategy when encountering a positive RT-PCR sample should be to reexamine the MRD levels in new samples obtained with a span of at least a fortnight, only designating the patient to be in MR if similar or increasing levels of the MRD marker in question are observed.
Clinical intervention based on MRD results: the critical issue
Outside clinical trials, it is still to a large extent a matter of personal taste how RT-PCR is incorporated into the patient workup, but given the known kinetics for the aberrations seen in AML and exemplified for the WT1+ patient in Figure 3, the literature is quite convincing in showing that patients turning PCR+ will not ever revert to an MRD− state.
The relationship between expression of an overexpressed MRD marker (eg, WT1) in PB and BM and MRD sensitivity in PB and BM. In the example in this figure, (BM MRD level/PB MRD level) = (BM physiologic level/PB physiologic level), making the 2 tissues equally useful for MRD detection (period between MR and HR equal).
The relationship between expression of an overexpressed MRD marker (eg, WT1) in PB and BM and MRD sensitivity in PB and BM. In the example in this figure, (BM MRD level/PB MRD level) = (BM physiologic level/PB physiologic level), making the 2 tissues equally useful for MRD detection (period between MR and HR equal).
What is then the evidence for a beneficial effect of MRD intervention at present? For PML-RARA+ patients, a beneficial effect of action on MR has been shown both by Italian31 and Spanish32 APL groups. Recently, preemptive treatment was shown to be feasible for PML-RARA+ patients monitored as part of the large AML15 trial.30
Likewise, a study, albeit in acute lymphoblastic leukemia, from the German Cooperative group has resulted in these patients being treated now according to MRD detection parameters (in casu Ig-receptor gene rearrangement status).33 In non-APL AML, a Czech group has published their results treating molecular relapses using anti-CD33, “5 + 2”-like chemotherapy, donor lymphocyte infusion, or discontinuation of immunosuppression, showing that administration of preemptive therapy can often postpone HR but that HR will ultimately occur in a large majority of cases (10 of 13 relapses treated).34 None of these studies entails a randomized comparison between action and no action on MR, and it must be admitted that no such studies are available to document the effect of preemptive action. However, a very stringently performed study by Rubnitz et al35 has, however, shown encouraging results in childhood AML, where risk-adapted treatment according to MRD criteria resulted in improved outcome. Together, these papers could be considered as proof of principle for MRD-based patient stratification. Although this is very important, it does not imply that a patient stratified to a certain therapy should be treated at MR.
Thus, a decision to intervene does not necessarily need to be standard cytoreduction. Wait-and-watch while commencing a donor search in patients eligible for allogeneic transplantation as well the administration of donor lymphocyte infusion36 in patients after allogeneic transplantation represents less drastic treatment options. Likewise, commencement of biologic therapy (eg, anti-CD33) or molecular targeting (eg, all-trans retinoic acid, tyrosine kinase inhibitors, or FLT3-ITD inhibitors) constitute equally valid, if as of yet not formally validated, alternatives with lower side effect profiles.30,34
The role of overexpressed genes in MRD detection
Although we think that the first question has already been resolved showing that, for example, WT1 can act as a bona fide marker and refer the reader to recent papers documenting this,36-38 an outstanding issue pertains to the fact that these genes are often expressed in normal BM, and in exceptional cases even in PB (this presents a special problem in pediatric AML patients with infections; Henrik Hasle, oral communication, October 2010). This will result in different criteria being applied to the interpretation of PB and BM separately (Figure 3). We think that extra caution should be exerted in WT1+ patients with possible MR. Thus, allowing for extra samples to be obtained from these patients seems judicious. Although data are strong that PB will suffice in most situations16,27,38,39 given that the occasional patient (in our hands, 2%-3%) will regenerate hematopoiesis after cytoreduction to a WT1 level above a given threshold level, liberal use should be granted to BM aspirates.
This strategy is illustrated in Figure 4, where a young AML patient was followed by quarterly PB sampling and, after several positive samples with increasing levels of WT1 after molecular conversion, BM sampled and subsequently referred to an allogeneic transplantation, where, notably, donor search had commenced at first PB conversion. Importantly, this patient did not experience a hematologic relapse.
Follow-up during therapy, CR, and relapse of a WT1+ patient treated at our department. x-axis represents time since diagnosis; y-axis represents WT1 expression shown as fraction of the expression at diagnosis.
Follow-up during therapy, CR, and relapse of a WT1+ patient treated at our department. x-axis represents time since diagnosis; y-axis represents WT1 expression shown as fraction of the expression at diagnosis.
Kinetics of relapse according to molecular aberration
From the literature, it is now safe to construct a kinetic hierarchy for RT-PCR conversion (ie, the timing of molecular relapse as a function of time to hematologic relapse). Somewhat surprisingly, given the overall favorable prognosis of these patients, those harboring PML-RARA fusion transcripts seem to be those who display the fastest relapses. This has been amply documented in a recent study from the AML15 team,27,30 who also addressed the important question of preemptive intervention in a smaller series of patients and found that this had a clear favorable effect.
The next surprising finding regarding relapse kinetics is that core-binding factor leukemias, generally considered to have similar biologic features, are strikingly different in relapse speed, with RUNX1-RUNX1T1 patients displaying a somewhat more sluggish pattern than PML-RARA patients on the one hand, but in no way as slow as CBFB-MYH11 patients on the other. In the latter, MR can indeed precede HR for up to one year,26,27,30 often with remarkably similar doubling times of the malignant clones between patients. The latter finding contrasts to the situation in NPM1c+ patients, where the rapidity of relapse observed in close longitudinal testing varies greatly.40 Reassuringly, with respect to the biologic relevance of molecular changes in AML, we recently observed in a multicenter study, with data largely from the Munich Leukemia Laboratory, that this heterogeneity was closely related to the FLT3 status, with patients harboring FLT3-ITD displaying the fastest relapses.27 For WT1, we found that, although being inferior to all other aberrations with the exception of PML-RARA, it is still a marker, which in theory provides a useful window of opportunity (Figure 4).27
Collectively, these data underline the importance of molecular testing in AML but also set the stage for a closer look at how and how often patients should be followed during CR. In this setting, it should be remembered that the initial molecular workup must be exhaustive enabling the opportunity for monitoring more than one marker during follow-up.
Frequency of MRD sampling
When MRD sampling was made possible in the late 1990s, the question quickly arose how often sampling was necessary to detect an impending relapse. Diverio et al24 conducted a pioneering study on PML-RARA+ leukemia and found that, using a 3-monthly sampling schedule, 20 of 28 relapses could be foreseen. The failures were explained by fast reappearance of the leukemic clone in 7 cases. In the last case, no sample had been received at the laboratory during the period preceding the relapse.
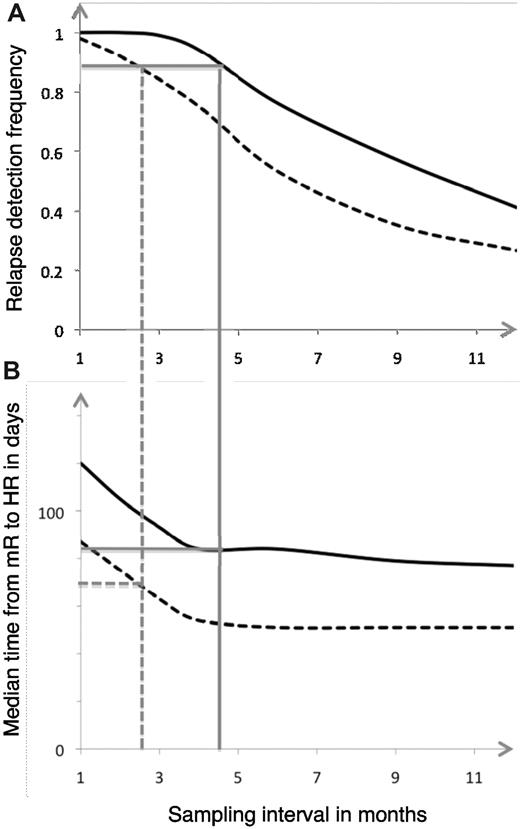
Several groups have since then repeated the finding that sampling intervals are of paramount importance to detect a high number of MRs before HR. Moreover, recent literature now allows us to more closely delineate how often the different molecular lesions should be assayed for to ensure timely MR detections. Here, we have taken the approach of analyzing AML CR patients who relapsed and in whom we had several MRD determinations before that. Realizing that patients were not followed by the same cadenzas of sampling, we took advantage of mathematical modeling to assess the relationship between sampling interval on the one hand and the likelihood of detection an impending relapse on the other. As detailed in Figure 5, such modeling can be made flexible so that it can be adapted to the wishes of the given center and the given molecular aberration. We consider this approach an advance in our handling of AML patients in CR because it will allow for the rational design of future trials, be they design using traditional cytoreduction or encompassing novel compounds targeted at the genetic lesions themselves, such as is the situation in chronic myeloid leukemia.
Relationship between MR detection and sampling interval. Correlation between relapse detection frequency and sampling interval (A) as well as median time from MR to HR in days, depending on sampling frequency (B) for RUNX1-RUNX1T1–based MRD follow-up. Full lines (BM) and broken lines (PB) represent sampling interval necessary to achieve 90% relapse detection. The corresponding median times from MR to HR can be seen in panel B.
Relationship between MR detection and sampling interval. Correlation between relapse detection frequency and sampling interval (A) as well as median time from MR to HR in days, depending on sampling frequency (B) for RUNX1-RUNX1T1–based MRD follow-up. Full lines (BM) and broken lines (PB) represent sampling interval necessary to achieve 90% relapse detection. The corresponding median times from MR to HR can be seen in panel B.
When will PB sampling suffice, and is BM material necessary?
After the introduction of the RT-PCR assay for MRD detection, it was initially assumed that PB sampling would suffice in the majority of situations. Although we now know that this viewpoint was too simplistic, the notion still holds true in for some of the aberrations discussed herein. Thus, from the relapse kinetics discussed above in conjunction with the limited number of direct PB versus BM comparisons at the time of MR in the literature, it is now safe to state that for the slowest relapses, such as CBFB-MYH11, NPM1c+ (the FLT3-ITD− ones), as well as RUNX1-RUNX1T1 PB sampling, will clearly suffice to ensure a window of opportunity on which to act. Whether to apply the same strategy for WT1+ patients will be a matter of opinion. We rarely perform BM biopsies, resorting to PB sampling quarterly as a means of MR detection. For the rare PML-RARA+ patients who will relapse, the European LeukemiaNet experience is that BM sampling will be necessary to detect a acceptable majority of these.27 For PB sampling, even with a strategy of monthly samplings, our relapse kinetic-based modeling suggests that only 75% of relapses will be detected with a median window-of opportunity before HR of only 35 days (Table 2).
Proposed guidelines for MRD follow-up using CBFB-MYH11, RUNX1-RUNX1T1, PML-RARA, NPM1c, or WT1 as molecular markers
| . | Sampling interval, mo . | RDF, % . | tm, d . |
|---|---|---|---|
| CBFB-MYH11 | |||
| PB | 6* | 90 | 180 |
| BM | Avoid | ||
| RUNX1-RUNX1T1 | |||
| PB | 3 | 85 | 65 |
| BM | 4 | 95 | 85 |
| PML-RARA | |||
| PB | 1 | 75 | 35 |
| BM | 2 | 90 | 60 |
| NPM1c/FLT3-ITD− | |||
| PB | 4 | 90 | 60 |
| BM | 6 | 90 | 120 |
| NPM1c/FLT3-ITD+ | |||
| PB | 3 | 90 | 65 |
| BM | 4 | 90 | 105 |
| WT1 | |||
| PB | 2 | 85 | 45 |
| BM | 4 | 95 | 75 |
| . | Sampling interval, mo . | RDF, % . | tm, d . |
|---|---|---|---|
| CBFB-MYH11 | |||
| PB | 6* | 90 | 180 |
| BM | Avoid | ||
| RUNX1-RUNX1T1 | |||
| PB | 3 | 85 | 65 |
| BM | 4 | 95 | 85 |
| PML-RARA | |||
| PB | 1 | 75 | 35 |
| BM | 2 | 90 | 60 |
| NPM1c/FLT3-ITD− | |||
| PB | 4 | 90 | 60 |
| BM | 6 | 90 | 120 |
| NPM1c/FLT3-ITD+ | |||
| PB | 3 | 90 | 65 |
| BM | 4 | 90 | 105 |
| WT1 | |||
| PB | 2 | 85 | 45 |
| BM | 4 | 95 | 75 |
RDF indicates relapse detection frequency; and tm, median time from molecular to hematologic relapse.
One additional MRD sampling recommended 3 months after end of chemotherapy to detect early relapses.
A proposal for guidelines for the application of MRD assays in AML patients in CR
Based on the data presented herein and the mathematical models developed previously,16,27 we think that a set of guidelines for MRD follow-up of the described MRD markers can be suggested. Although the models are flexible according to detection goals with respect to relapse detection frequencies (RDFs) and median times from MR to HR (tms), the data in Table 2 are based on the detection of 85% of relapses with a minimal tm of 60 days. As shown in Table 2, for PML-RARA, shorter tms were accepted given that salvage therapy in these patients does not usually involve an allogeneic stem cell transplantation. For other lesions, sampling every third month as the most frequent cadenza with the avoidance of BM sampling and on half-yearly PB sampling for CBFB-MYH11+ patients. The exception from this rule is WT1, where PB sampling every second month seems prudent.
The issue as to when to stop MRD monitoring has important implications, not the least of which is the avoidance of sampling patients, who are unlikely to ever experience a relapse. From historical data,41 we suggest stopping MRD surveillance after 3 years. This is in concordance with reported observations that by far the most patients occur before the 2-year point after diagnosis, thus allowing for a few extra MRD determinations in patients who have already been accustomed to the hassle of MRD testing. This might, to some extent, pertain to the issue of invoked costs for the samplings, but these must be considered to be negligible compared with the total ones inferred through cytoreduction and treatment of side effects. Finally, one needs to consider the logistics with respect to handling and shipment of samples. Here, it should be remembered that different fusion transcripts degrade differently12 and that care should be taken to immediately stabilize samples in suitable tubes. We would, at present, advocate shipping samples to laboratories with documented expertise in the various aspects of the techniques. On the other hand, given the preclinical standardization of the assay,7 decentralization to laboratories servicing at least 1.0 million inhabitants (corresponding to at least 500 assays/year) would be feasible in the not so distant future.
Future directions
As outlined in the Introduction, the genetic background for AML is diverse, and taking also epigenetic alterations into account, probably no 2 AML patients are identical with respect to their molecular phenotype. Naturally, this notion has resulted in ongoing effort to characterize AML subtypes with the aim to optimize pretherapeutic prognostication.2,42 In this setting, validated and optimized intratreatment MRD testing techniques offer the opportunity to incorporate the response to cytoreduction of the leukemic clone in the overall patient relapse risk assessment. Moreover, with optimized post-therapy sequential MRD follow-up (eg, according to guidelines like those suggested in “A proposal for guidelines for the application of MRD assays in AML patients in CR”), a patient-specific approach could be in sight. Although the concept of individualized therapy will hopefully also encompass such parameters as upfront molecular phenotyping, the vision of individualized patient treatment should include personalized intensification therapy, preferably based on precise chemoresistance characterization of the leukemic clone. In this setting, consolidation adapted therapy of CR patients administered according to longitudinal MRD measurements could well result in a quantum leap in the survival of these patients.
Although we strongly believe in preemptive therapy at MR in AML, we also realize that the present data are still not mature enough to warrant the individualized approach to which we allude. How should studies be designed, which will hopefully provide the sorely needed hard data? Such should first encompass as many markers as possible. This is by no means an insurmountable task because the 5 assays described here will provide more than 80% of AML patients with an MRD “handle.” Second, the study should be powered to yield results as early as possible (ie, with a rapid accrual rate, as in the United Kingdom-AML studies). Third, the design will need to be strictly randomized, meaning that participating centers not used to sample and analyze for MRD by RT-PCR will need to ship to other centers. Once more, the United Kingdom-AML trials seem to be the most promising in yielding definitive responses to these important questions. Thus, it seems the within the Working Parties on Leukemia AML17 trial a validation process for sampling logistics has nearly been completed and a “monitor versus no-monitor” randomization is planned to open in 2011 (Alan Burnett, e-mail communication, November 2010). Here, clinicians used to prescribing and reviewing MRD reports will in half of their patients, need to dispense of these. Only by this approach will it be assured that the far too long period of time during which the highly validated RT-PCR methodology has been in the wings can be brought to an end by it being brought to center stage for the benefit of the patient.
Acknowledgments
The authors thank our patients for contributing samples, and for continuous input during these efforts. H.B.O. thanks Drs David Grimwade and Susanne Schnittger for valuable contributions during the development of the mathematical models.
P.H. was supported by the Danish Cancer Society, the Danish MRC, the Danish Childhood Cancer Foundation, and the Karen Elise Jensen Foundation. Important parts of this work was carried out as part of the work package 12 initiative chaired by Dr Grimwade under the European Leukemia Network cooperative network.
Authorship
Contribution: P.H. wrote the first draft of the paper; H.B.O. contributed to the writing and editing and prepared the tables and figures; and P.H. and H.B.O. finalized the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Peter Hokland, Århus Sygehus, Tage-Hansens Gade 2, 8000 Århus-C, DK Denmark; e-mail: phokland@ki.au.dk.