Abstract
The mechanisms of resistance to tyrosine kinase inhibitors (TKIs) in chronic myeloid leukemia (CML) often remain obscure. Analysis of patient samples during disease progression revealed the up-regulation of the oncogene TWIST-1, also measured in primary samples from TKI-resistant patients. Moreover, we found that TWIST-1 was overexpressed in CML diagnostic samples of patients who later developed cytogenetic resistance to imatinib, even those without any detectable resistance mechanism. We confirmed the up-regulation of TWIST-1 at both RNA and protein levels in imatinib-resistant cell lines, irrespective of any other resistance mechanism. Analysis with specific small interfering RNA suggested TWIST-1 involvement in the resistance phenotype. Finally, the kinetics of TWIST-1 expression during the individual medical histories of CML patients indicated that TWIST-1 expression is down-regulated by TKIs and up-regulated with TKI resistance. We hypothesize that the overexpression of the TWIST-1 oncogene represents a novel key prognostic factor potentially useful for optimizing CML management in the TKI era.
Introduction
Chronic myeloid leukemia (CML) is a model for cancer stem cell and tyrosine kinase inhibitor (TKI) resistance. If chronic phase (CP) patients are discriminated by Sokal score1 at diagnosis, they remain heterogeneous and lack a reliable molecular prognosis marker. Imatinib has become the standard of CP-CML care,2 but some rare Philadelphia-positive (Ph+) stem cells may be insensitive to TKIs,3-5 maintain detectable disease over years, and represent a reservoir in which secondary molecular events occur and initiate disease progression. The BMI-1 gene, involved in stem cell self-renewal and maintenance, has been proposed to predict CML disease progression.6 We hypothesized that TWIST-1, a transcription factor involved in embryogenesis, undifferentiated cell proliferation, and cell survival,7-9 might be involved in CML. We analyzed its expression in CML cells from patients and in cell lines and identified it as a unique predictive marker for TKI resistance, especially in patients with no other detectable resistance mechanism.
Methods
After informed consent in accordance with the Declaration of Helsinki and local ethics committee bylaws (from the Délégation à la recherche clinique des Hospices Civils de Lyon, Lyon, France), samples were obtained from CML patients at diagnosis or during follow-up and from healthy allogeneic donors. CML cell lines were cultured as described.10 CD34-purified cells (StemCell Technologies) were placed in Tri-Reagent (Sigma-Aldrich). Total RNAs were reverse-transcribed, and quantitative polymerase chain reaction (PCR) was performed using sequence-specific primers (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article; LightCycler480II system, Roche Diagnostics and QuantiFAST SYBR Green I-kit, QIAGEN) using a fast protocol (activation, 5 minutes at 95°C; amplification,10 seconds at 95°C, 30 seconds at 60°C; melting, 10 seconds at 95°C, 10 seconds at 50°C). Specific TWIST-small interfering RNA (siRNA; sequence coding regions 55-75) or a Scramble-negative control were transfected as duplexes (RNAiMax, Invitrogen).7-9 Cells were counted (trypan blue staining), controlled for transfer efficiency (flow cytometry BDFACSCalibur), or analyzed for TWIST-1 silencing (RNA and protein). Western blot was performed as described11 using monoclonal antibodies to ABL (BD Biosciences PharMingen), TWIST-1 (Abnova), and Ku80 (Roche Diagnostics). Statistically, the Wilcoxon Mann-Whitney test, with R Version 2.9.2 software, assessed differences between CML phases, responding and nonresponding patients or CD34+ and CD34− fractions, and the Student paired t test for differences between cell lines groups. In addition, we performed an analysis of variance applying a bivariate analysis. Significant P values (P < .05) are given in the text, supplemental Table 2, and figures.
Results and discussion
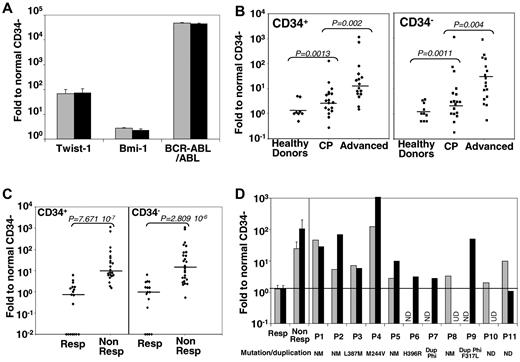
Approximately 30% of CP-CML patients develop resistance to TKIs, the mechanism(s) of which remain(s) obscure. Genome-wide microarray analysis of CML cell lines has shown that TWIST-1, elevated in other cancers, immature cells, and patients with drug resistance,12 is deregulated in a CML-resistant cell line.13 We analyzed TWIST-1 expression by quantitative PCR in purified immature (CD34+) and mature (CD34−) cells from patients at different disease phases and after emergence of TKI resistance. Compared with normal samples, the overall TWIST-1, BMI-1, and BCR-ABL expressions increased (Figure 1A) in CD34+ and CD34− cells of, respectively 66-fold (P < .001) and 74-fold (P < .001) for TWIST-1 and 2.71-fold (P < .001) and 2.25-fold (P < .001) for BMI-1 as reported.6 Along disease progression, TWIST-1 expression significantly increased in CD34+ cells (11-fold and up to 133-fold in chronic and advanced phases, respectively; Figure 1B) and CD34− cells (65-fold and 97-fold). Comparing TKI responder and nonresponder samples, we observed an approximately 100-fold increase in TWIST-1 expression, regardless of CD34 expression (P = .88; Figure 1C), strongly supporting its expression as a predictive factor for CML response to TKIs, unlike BMI-1 (supplemental Figures 1A, 2B). In accordance with previous data, we observed an increased expression of BCR-ABL between CD34+ and CD34− cells (supplemental Figure 1A-B). We analyzed TWIST-1 expression in CML samples at diagnosis in a series of samples from 32 patients and observed an increased mean value at diagnosis for nonresponders (24.65-fold and 107.44-fold) compared with responders (1.28-fold and 1.32-fold), respectively, for CD34+ and CD34− cells. Indeed, sensitive patients, before treatment (Figure 1D mean responder) or during treatment (Figure 1C responder), always presented a level of TWIST-1 similar to that of normal donors. The individual values of all patients (P1-P11) who developed resistance (European LeukemiaNet criteria14 ) later in their disease course had higher TWIST-1 expression than the mean value of responding patients (Figure 1D; supplemental Table 2), whereas only 2 had increased BMI-1 expression (supplemental Figure 1C). Subsequently, we think that TWIST-1 represents a unique and reliable predictive marker of TKI resistance in CML regardless of the resistance origin (Figure 1D; supplemental Table 2). Compared with the responder mean values, all nonresponder patient samples displayed a significant increased expression of TWIST-1 in CD34+ cells (100% of nonresponder patients), whereas only 8 of 11 tested patients (72% of nonresponder patients) had an increased Twist-1 expression in their CD34− compartment (supplemental Table 2). Nevertheless, the difference between responder and nonresponder values of Twist-1 in CD34− remained statistically significant (supplemental Table 2). This was confirmed by the detection of TWIST-1 expression within mononuclear cells from those patients plus one in accelerated phase and one in myeloid blast crisis (supplemental Table 3). In addition, sorted CD19+ cells from the patient 10 sample at diagnosis were negative for TWIST-1 expression despite a BCR-ABL positivity (not shown). In addition, from the 21 TKI-responsive tested patients at diagnosis, only 3 and 4 slightly expressed TWIST-1 more than 2-fold, in their CD34+ and CD34− compartments, respectively (supplemental Table 2). Therefore, TWIST-1 measurement in CD34+ cells appears to be the most reliable parameter to discriminate since diagnosis of imatinib responders or nonresponders.
TWIST-1 expression in primary CD34+ and CD34− selected CML cells. Gene expression in CD34 immuno-selected hematopoietic cells from CML patients. Results are expressed as a ratio to normal (healthy donors) CD34− cell values. (A) Comparative expression of TWIST-1, BMI-1, and BCR-ABL in CD34+ ( ) and CD34− (■) compartments from CML samples (n = 40). (B) Comparative expression of TWIST-1 in CD34+ (left panel) and CD34− (right panel) cells in controls (healthy BM donors; n = 10), in CP (n = 20), and in advanced-phase (n = 16) CML cells. Horizontal bars represent the medians. (C) Comparative expression of TWIST-1, in CD34+ (left panel) and CD34− (right panel) cells in CML patients responsive (Resp; n = 18) or nonresponsive (Non Resp; n = 25) to imatinib. Horizontal bars represent the medians. (D) Mean TWIST-1 expression in CD34+ (
) and CD34− (■) compartments from CML samples (n = 40). (B) Comparative expression of TWIST-1 in CD34+ (left panel) and CD34− (right panel) cells in controls (healthy BM donors; n = 10), in CP (n = 20), and in advanced-phase (n = 16) CML cells. Horizontal bars represent the medians. (C) Comparative expression of TWIST-1, in CD34+ (left panel) and CD34− (right panel) cells in CML patients responsive (Resp; n = 18) or nonresponsive (Non Resp; n = 25) to imatinib. Horizontal bars represent the medians. (D) Mean TWIST-1 expression in CD34+ ( ) or CD34− (■) CML cells from a total of 32 patients at diagnosis before any treatment. P1 to P11 indicate individual data from patient 1 (P1) to patient 11 (P11). Their mutational status (NM indicates nonmutated; and M, mutated) or presence of chromosome Ph duplication (Dup Phi) status are indicated below the graph. ND indicates not done; and UD, undetected.
) or CD34− (■) CML cells from a total of 32 patients at diagnosis before any treatment. P1 to P11 indicate individual data from patient 1 (P1) to patient 11 (P11). Their mutational status (NM indicates nonmutated; and M, mutated) or presence of chromosome Ph duplication (Dup Phi) status are indicated below the graph. ND indicates not done; and UD, undetected.
TWIST-1 expression in primary CD34+ and CD34− selected CML cells. Gene expression in CD34 immuno-selected hematopoietic cells from CML patients. Results are expressed as a ratio to normal (healthy donors) CD34− cell values. (A) Comparative expression of TWIST-1, BMI-1, and BCR-ABL in CD34+ ( ) and CD34− (■) compartments from CML samples (n = 40). (B) Comparative expression of TWIST-1 in CD34+ (left panel) and CD34− (right panel) cells in controls (healthy BM donors; n = 10), in CP (n = 20), and in advanced-phase (n = 16) CML cells. Horizontal bars represent the medians. (C) Comparative expression of TWIST-1, in CD34+ (left panel) and CD34− (right panel) cells in CML patients responsive (Resp; n = 18) or nonresponsive (Non Resp; n = 25) to imatinib. Horizontal bars represent the medians. (D) Mean TWIST-1 expression in CD34+ (
) and CD34− (■) compartments from CML samples (n = 40). (B) Comparative expression of TWIST-1 in CD34+ (left panel) and CD34− (right panel) cells in controls (healthy BM donors; n = 10), in CP (n = 20), and in advanced-phase (n = 16) CML cells. Horizontal bars represent the medians. (C) Comparative expression of TWIST-1, in CD34+ (left panel) and CD34− (right panel) cells in CML patients responsive (Resp; n = 18) or nonresponsive (Non Resp; n = 25) to imatinib. Horizontal bars represent the medians. (D) Mean TWIST-1 expression in CD34+ ( ) or CD34− (■) CML cells from a total of 32 patients at diagnosis before any treatment. P1 to P11 indicate individual data from patient 1 (P1) to patient 11 (P11). Their mutational status (NM indicates nonmutated; and M, mutated) or presence of chromosome Ph duplication (Dup Phi) status are indicated below the graph. ND indicates not done; and UD, undetected.
) or CD34− (■) CML cells from a total of 32 patients at diagnosis before any treatment. P1 to P11 indicate individual data from patient 1 (P1) to patient 11 (P11). Their mutational status (NM indicates nonmutated; and M, mutated) or presence of chromosome Ph duplication (Dup Phi) status are indicated below the graph. ND indicates not done; and UD, undetected.
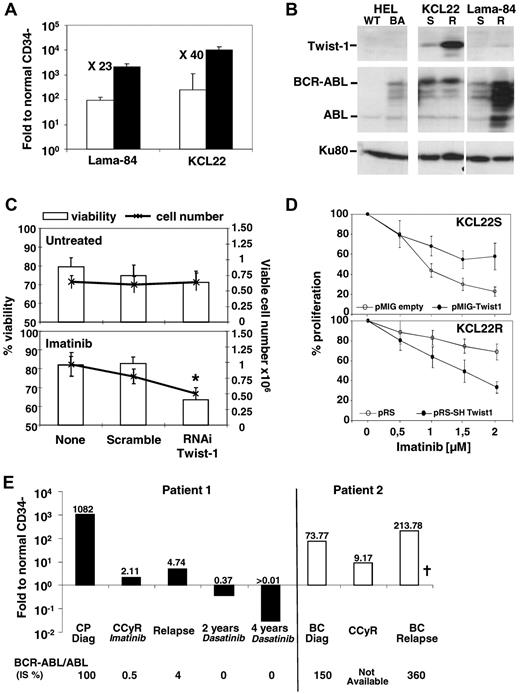
Our data obtained in primary CML samples confirmed TWIST-1 deregulation in the KCL22 CML cell line13 (resistant through unknown mechanisms). Quantitative PCR showed a 23- and 40-fold TWIST-1 expression increase (Figure 2A), with resistance in Lama-84 and KCL22, respectively, and confirmed at protein level (Figure 2B). Interestingly, TWIST-1 overexpression seems to be independent from BCR-ABL protein levels as observed in HEL, KCL22R, and Lama-84R cells10 (Figure 2B). TWIST-1 silencing by RNAi in Lama-84R and KCL22R (supplemental Figure 2) cells had no effect by itself but decreased cell numbers and viability in response to imatinib (Figure 2C-D). Conversely, overexpression of TWIST-1 in KCL22-sensitive cells (supplemental Figure 2B) significantly improved their resistance to increasing doses of imatinib (Figure 2D). Altogether, these data indicate that TWIST-1 expression is involved in imatinib resistance. Finally, we analyzed TWIST-1 expression in CD34− cells during disease progression; and although it was highly expressed at diagnosis, its expression significantly decreased after complete cytogenetic remission (Figure 2E) and increased again at relapse, unlike BMI-1 (supplemental Figure 3).
Involvement of TWIST-1 in CML response to TKIs, KCL22, or Lama-84 cell lines was analyzed for TWIST-1 expression. (A) Comparative expression by quantitative PCR of TWIST-1 in Lama-84 and KCL-22 imatinib-sensitive (□) and imatinib-resistant (■) Ph1+ cell lines. Results are expressed as a ratio to normal (healthy donors) CD34− cell values. (B) TWIST-1, BCR-ABL, and KU80 (used as loading control) protein analysis by Western blot in HEL expressing BCR-ABL (BA) or not (WT), KCL-22 and Lama 84 imatinib-sensitive (S) and -resistant (R) cell extracts. (C) Viability and viable cell numbers in untreated and imatinib-treated (1.5μM) Lama84-resistant Ph1+ cell line, submitted to TWIST-1 siRNA or negative scramble as control. Cells were incubated in 96-well plates in serum-free medium with stem cell factor (100 ng/mL), in the presence of 50nM of fluorescent siRNA. Results are presented as a percentage of viable cells or as total viable cell numbers ± SEM. After 2 days of culture, cell proliferation and viability were determined by trypan blue counting. Fold proliferation was determined by reference to the input number of viable cells. Results represent the mean ± SEM of 7 independent experiments. *Significant value (P < .05). (D) TWIST-1 expression in KCL22-resistant cells (KCL22R) transduced with a pSR vector empty or containing sh sequence against TWIST-1 or in KCl22-sensitive cells (KCL22S) transduced with a pMIG vector empty or containing TWIST-1 expressing sequence. After 3 days in the presence of puromycin selection for KCL22R-shTWIST-1 or after 48 hours for KCL22S-TWIST-1, cells were incubated in the presence of imatinib at the indicated dose. After 3 days of culture, cell proliferation and viability were determined by trypan blue counting. The percentage of proliferation was determined by reference to the nontreated number of viable cells ± SEM from 5 experiments. (E) Follow-up analysis for TWIST-1 expression by quantitative PCR in the CD34− fraction. Results are expressed as a ratio to normal (healthy donors) CD34− cell values. The values obtained for the BCR-ABL/ABL ratio are indicated as a percentage (IS) at each data point for both patients. BC indicates blast crisis; and CCyR, complete cytogenetic remission. †Deceased.
Involvement of TWIST-1 in CML response to TKIs, KCL22, or Lama-84 cell lines was analyzed for TWIST-1 expression. (A) Comparative expression by quantitative PCR of TWIST-1 in Lama-84 and KCL-22 imatinib-sensitive (□) and imatinib-resistant (■) Ph1+ cell lines. Results are expressed as a ratio to normal (healthy donors) CD34− cell values. (B) TWIST-1, BCR-ABL, and KU80 (used as loading control) protein analysis by Western blot in HEL expressing BCR-ABL (BA) or not (WT), KCL-22 and Lama 84 imatinib-sensitive (S) and -resistant (R) cell extracts. (C) Viability and viable cell numbers in untreated and imatinib-treated (1.5μM) Lama84-resistant Ph1+ cell line, submitted to TWIST-1 siRNA or negative scramble as control. Cells were incubated in 96-well plates in serum-free medium with stem cell factor (100 ng/mL), in the presence of 50nM of fluorescent siRNA. Results are presented as a percentage of viable cells or as total viable cell numbers ± SEM. After 2 days of culture, cell proliferation and viability were determined by trypan blue counting. Fold proliferation was determined by reference to the input number of viable cells. Results represent the mean ± SEM of 7 independent experiments. *Significant value (P < .05). (D) TWIST-1 expression in KCL22-resistant cells (KCL22R) transduced with a pSR vector empty or containing sh sequence against TWIST-1 or in KCl22-sensitive cells (KCL22S) transduced with a pMIG vector empty or containing TWIST-1 expressing sequence. After 3 days in the presence of puromycin selection for KCL22R-shTWIST-1 or after 48 hours for KCL22S-TWIST-1, cells were incubated in the presence of imatinib at the indicated dose. After 3 days of culture, cell proliferation and viability were determined by trypan blue counting. The percentage of proliferation was determined by reference to the nontreated number of viable cells ± SEM from 5 experiments. (E) Follow-up analysis for TWIST-1 expression by quantitative PCR in the CD34− fraction. Results are expressed as a ratio to normal (healthy donors) CD34− cell values. The values obtained for the BCR-ABL/ABL ratio are indicated as a percentage (IS) at each data point for both patients. BC indicates blast crisis; and CCyR, complete cytogenetic remission. †Deceased.
In conclusion, this study demonstrates that TWIST-1 is deregulated in CML patients at diagnosis and during progression and in cells innately imatinib-resistant through a yet unknown mechanism. Our results indicate that, with BCR-ABL monitoring, TWIST-1 could represent a powerful biomarker for early detection of TKI resistance at diagnosis and during residual disease follow-up. TWIST-1 might act as a survival factor providing a selective growth advantage contributing to drug resistance, as reported in solid tumors.12,15-17 TKI-resistant TWIST-1–overexpressing cells might emerge from leukemic Ph+ stem cells, but further work is necessary to evaluate the link with BCR-ABL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank S. Joly (U590), K. Chabane (Centre Hospitalier Lyon Sud), and E. Gadolet (Hematology department, Hôpital Edouard Herriot) for technical assistance and S. Morisset for statistical analysis (Association ARCHE, Hôpital Edouard Herriot).
This work was supported in part by Inserm (V.M.-S., A.P.) and Novartis Pharma France (K.S.).
Authorship
Contribution: E.C., G.H., S.J., and K.S. performed experiments and analyzed data; S.H. provided routine laboratory clinical data following international guidelines; T.V. contributed to research design and analyzed data; F.-X.M. provided cell lines and proofread the manuscript; C.D. contributed to discussions and proofread the manuscript; A.P. provided TWIST-1-vectors and proofread the manuscript; F.E.N. followed up the patients, provided primary cells, analyzed the data, and wrote the paper; and V.M.-S. performed research design, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Véronique Maguer-Satta, Inserm Unité U590, Centre Léon Bérard, 28 Rue Laennec, 69373 Lyon Cedex 08, France; e-mail: maguer@lyon.fnclcc.fr; and Franck Nicolini, Hospices Civils de Lyon, Hematology department, E. Herriot Hospital, 5 place d'Arsonval, 69437, Lyon, France; e-mail: franck-emmanuel.nicolini@chu-lyon.fr.
References
Author notes
E.C. and G.H. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal