Abstract
Congenital dyserythropoietic anemia type 1 (CDA-1), a rare inborn anemia characterized by abnormal chromatin ultrastructure in erythroblasts, is caused by abnormalities in codanin-1, a highly conserved protein of unknown function. We have produced 3 monoclonal antibodies to codanin-1 that demonstrate its distribution in both nucleus and cytoplasm by immunofluorescence and allow quantitative measurements of patient and normal material by Western blot. A detailed analysis of chromatin structure in CDA-1 erythroblasts shows no abnormalities in overall histone composition, and the genome-wide epigenetic landscape of several histone modifications is maintained. However, immunofluorescence analysis of intermediate erythroblasts from patients with CDA-1 reveals abnormal accumulation of HP1α in the Golgi apparatus. A link between mutant codanin-1 and the aberrant localization of HP1α is supported by the finding that codanin-1 can be coimmunoprecipitated by anti-HP1α antibodies. Furthermore, we show colocalization of codanin-1 with Sec23B, the protein defective in CDA-2 suggesting that the CDAs might be linked at the molecular level. Mice containing a gene-trapped Cdan1 locus demonstrate its widespread expression during development. Cdan1gt/gt homozygotes die in utero before the onset of primitive erythropoiesis, suggesting that Cdan1 has other critical roles during embryogenesis.
Introduction
The congenital dyserythropoietic anemias (CDAs) are a heterogeneous group of rare inborn disorders mainly affecting erythropoiesis.1 Distinct from other inherited bone marrow failure syndromes, they are marked by morphologic abnormalities of the erythroblasts that lead to ineffective erythropoiesis/dyserythropoiesis, whereas the other hematopoietic lineages appear to be unaffected. Based largely on the dysplastic changes observed in erythroblasts by light and electron microscopy, the CDAs have been divided into 3 major types, designated CDA types 1, 2, and 32 ; causative genes have been identified for CDA-1 (CDAN1/codanin-1)3 and CDA-2 (CDAN2/Sec23B),4 whereas only the chromosomal locus is known for CDA-3 (15q21–25).5
In CDA-1, the majority of cases come to attention during childhood and adolescence, often within episodes of erythropoietic stress (ie, infection, and in young women, pregnancy).6,7 Severe presentations can include gallstones, chronic hyperbilirubinemia, and/or extramedullary hematopoietic foci in the parietal and frontal bones of the skull (as observed in thalassemia). Although iron overloading is rarely the revelatory sign, it seems to be present in the majority of patients after childhood. In some cases, skeletal or other dysmorphic features can be identified, including short stature, distal limb and nail malformations, vertebral deformations/dysplasias, and skin pigmentation defects.1
The anemia is macrocytic with mean corpuscular volumes up to 120 fL, and the blood smear shows anisopoikilocytosis, basophilic stippling, and an inadequate reticulocyte response compared with hemolytic anemias. Light microscopy of the bone marrow shows erythroid hyperplasia with abnormal precursors displaying a megaloblastoid appearance. Dysplastic signs include markedly irregular nuclei with frequent (3.5%-7%) binucleate erythroblasts (particularly late cells) and occasional trinucleate and tetranucleate cells.8 None of these morphologic abnormalities is entirely specific for CDA-1 as they can be found in the other CDA types and in other acquired dyserythropoietic states.9 Particular diagnostic findings in CDA-1 are the presence in the marrow of internuclear bridges between intermediate erythroblasts (∼ 3% of total erythroblasts) and a characteristic pattern of spongy “Swiss cheese” heterochromatin as observed by electron microscopy. The pattern of inheritance is autosomal recessive, and obligate heterozygotes have normal hematologic indices, peripheral blood, and bone marrow morphology.
A large majority of CDA-1 cases are associated with missense mutations in the CDAN1 gene on chromosome 15q15, leading to single amino acid substitutions in the 134-kDa codanin-1 protein.3 A completely null genotype has never been identified in a CDA-1 patient, although rare non-sense mutations can be found in a heterozygous state. Codanin-1 has been evolutionarily highly conserved but its function is still unknown because it carries no previously described peptide motif or domain with similarity to any other within the existing eukaryotic proteome. Therefore, we examined the consequences of its mutation on chromatin structure in the erythroblasts of CDA-1 patients, and assessed the functional role of Cdan1 in a mouse model.
Methods
Patient material, cells, and culturing
Subjects were identified through physician-initiated referral and were included in the study based on diagnostic criteria previously described for CDA-1.7 Bone marrow and/or peripheral blood were obtained with informed consent in accordance with the Declaration of Helsinki and after institutional review board approval and Home Office approval.
Human erythroblasts, from patients or normal controls, were cultured from mononuclear cells (isolated from peripheral blood or buffy coat samples [National Blood Center, Bristol, United Kingdom]) in a 2-phase system described by Fibach et al.10 Erythroid development was monitored daily in the second phase of culturing by histologic and flow cytometric analyses (by CD71/transferrin receptor and glycophorin A staining) as outlined by Brown et al.11 Polychromatophilic erythroblasts (from day 8 or 9 of the second phase of culture) were defined as intermediate erythroblasts. Primary murine splenic erythroblasts were obtained from phenylhydrazine-treated adult mice, as described by Spivak et al.12 K562, HeLa, MEG-1, MEL, and GMMC cell lines were grown in RPMI 1640 medium (Sigma-Aldrich), 50 U/mL penicillin G (Invitrogen), 50 μg/mL streptomycin (Invitrogen), and 2mM l-glutamine (Invitrogen) supplemented with 10% (volume/volume) FCS (PAA Laboratories). Epstein-Barr virus (EBV)-transformed lymphoblastoid cell lines were obtained by immortalizing human peripheral blood mononuclear cells with EBV virus from healthy subjects and CDA-1 patients.
Antibody generation
Sequences from CDAN1 cDNA (obtained from pOTB7 vector; BGDP) encoding large protein fragments (amino acids 294-607 of codanin-1) were cloned into pOPINF vectors, which add a hexahistidine tag to the N-terminus of the protein, as described by Berrow et al.13 The protein was produced in inclusion bodies, solubilized in 50mM Tris, pH 8.5, 8M urea, and 30mM imidazole, and purified by nickel affinity chromatography. Purified protein was concentrated and buffer exchanged into 50mM Tris, pH 8.5, and 2M urea, using a centrifugal concentrator (Sartorius). MF1 mice were immunized twice subcutaneously with 30 μg of recombinant codanin-1 protein fragments emulsified in TiterMax Gold adjuvant (CytRx) at 10- to 14-day intervals. The mice received 2 booster inoculations in phosphate-buffered saline (PBS) intraperitoneally at 10- to 14-day intervals. On day 4, after the final booster, spleen cells were isolated and fused with NS-0 mouse myeloma cells in 50% polyethylene glycol 1500. Hybridomas secreting monoclonal antibodies against the recombinant protein fragments were screened by enzyme-linked immunosorbent assay and staining on COS-1 transfectants. Selected hybridomas were cloned by limiting dilution.
Antibodies
See supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Immunofluorescence and confocal microscopy
Cells were allowed to settle on poly-L-lysine–treated coverslips for 10 minutes. Cells were then fixed in 4% paraformaldehyde (Electron Microscope Services) for 15 minutes at room temperature and permeabilized in 1% Triton X-100 in PBS for 10 minutes at room temperature. Nonspecific binding sites were blocked using 10% FCS (PAA Laboratories) in PBS for 30 minutes at room temperature. Cells were stained with primary and secondary antibodies in block as listed in supplemental Table 1, followed by DNA counterstaining with TOPRO-3 (Invitrogen). Coverslips were mounted in Vectashield (Vector Laboratories) with 1 μg/mL 4′,6-diamidino-2-phenylindole (Roche Diagnostics) added. Analysis of fluorescent preparations was undertaken using a Bio-Rad Radiance 2000 confocal system (Carl Zeiss) on a BX51 upright microscope (Olympus). All images were collected using a 100×NA 1.3 UPlan F1 oil immersion objective (Olympus) at room temperature using LaserSharp software (Carl Zeiss).
X-Gal staining and brightfield microscopy
β-Galactosidase (X-Gal) staining of cultured cells and of murine embryos was performed after fixation and exposure to X-Gal as previously described.14 TOPRO-3 (Invitrogen) was used as a DNA counterstain. X-Gal–stained preparations were imaged on a BX60 microscope (Olympus) with a QI CAM CCD camera (QImaging) and OpenLab Version 4.0 (Improvision) software.
Electron microscopy
Cell suspensions were fixed in 4% glutaraldehyde in 0.1M phosphate buffer, postfixed in 2% osmium tetroxide in PBS, dehydrated in ethanol, and followed by immersion in propylene oxide before embedding in Spurr epoxy resin; 1-μm sections, stained with Azure A, were examined by light microscopy to identify areas of interest. Thin sections of suitable areas were cut and stained with uranyl acetate and lead citrate before examination on a 1200EX electron microscope (JEOL).
Protein isolation, SDS-PAGE, immunoblotting, and coimmunoprecipitation
Whole cell proteins were extracted by addition of lysis buffer (50mM Tris-HCl, pH 7.4, 0.5% NP-40 [volume/volume], 150mM NaCl, 20mM MgCl2, 1× protease inhibitors mini-ethylenediaminetetraacetic acid [Roche Diagnostics]). To obtain nuclear or cytosolic fractions, a hypotonic/hypertonic buffer separation technique was used.15 Nuclear, histone, and heterochromatin proteins were extracted by incubation in lysis buffer (0.2% Triton X-100 [volume/volume], 10mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.6, 1.5mM MgCl2, 10mM KCl, 10mM sodium butyrate, 1× protease inhibitors [Roche Diagnostics], 1× phosphatase inhibitors [Roche Diagnostics] in high performance liquid chromatography-grade water), followed by nuclear lysis in 150 μL 0.4M HCl. Nuclear membranes were removed by centrifugation, and supernatant proteins were precipitated with ice-cold acetone, followed by methanol and chloroform extraction. Proteins from all extraction methods were quantified by DC Protein Assay (Bio-Rad) and with a QuBit platform (Invitrogen).
Protein samples were separated by SDS-PAGE on NuPAGE/Novex gels (Invitrogen) in 2-N-morpholino-ethanesulfonic acid or 3-N-morpholino-propanesulfonic acid buffer with added antioxidants. Proteins were transferred by immunoblotting onto polyvinylidene difluoride membrane (Millipore), with selected pore size (0.2 μm for low molecular weight, 0.45 μm for intermediate to high molecular weight), using an XCell-II Blot module (Invitrogen). Nonspecific binding sites were blocked using 5% powdered milk and 0.2% Tween-20 (volume/volume) in PBS. Blots were then incubated with primary and secondary antibodies (described in supplemental Table 1) following the manufacturer's instructions, followed by horseradish peroxidase detection using the ECL Plus detection horseradish peroxidase kit (GE Healthcare Life Sciences). For coimmunoprecipitation of proteins, a Universal Magnetic coIP kit (Active Motif) was used. Proteins were precipitated using chloroform-methanol and resuspended in 2× Laemmli buffer before immunoblotting as described.16
ChIP-chip analysis
Antibodies used are described in supplemental Table 1. ChIP-chip analysis was performed as previously described by De Gobbi et al.17 Sequences were aligned onto the hg17 assembly using the UCSC Genome Browser (www.genome.ucsc.edu).18
DNA isolation, PCR, and sequencing
DNA was obtained by lysing cells in 10mM Tris-HCl, pH 8.0, 10mM NaCl, 10mM ethylenediaminetetraacetic acid, 0.5% SDS (volume/volume) and 50 g/mL proteinase K (Fermentas International) at a dilution of 5 × 106 cells/mL of buffer at 55°C for 3 hours, followed by phenol-chloroform extraction. DNA concentrations were determined using a Nanodrop spectrophotometer (Thermo Fisher Scientific).
PCR reactions were performed using Fermentas products (dNTP mix [each 0.25mM], 1× PCR reaction buffer, Taq DNA polymerase [1-5 U]) with 50 to 100 ng DNA, 1μM primers (Sigma-Aldrich), and PCR-grade H2O. Standard cycling conditions were as follows: 94°C at 10 minutes, 35 cycles of 30 seconds at 94°C, 45 seconds at 65°C, 60 seconds at 72°C, and a final 60 seconds at 72°C.
Sequencing of DNA fragments was performed using an ABI PRISM BigDye Terminator Version 3.1 Cycle Sequencing Kit and the ABI PRISM 3100 Genetic Analyzer (both Applied Biosystems). Sequences were analyzed using the ABI Prism and the 4Peaks Version 1.7.2 (Mekentosj BV) software suites.
RNA isolation, cDNA synthesis, RT-PCR, and quantitative PCR
Total RNA was isolated by TRIzol treatment (Invitrogen) and quantified on a BioAnalyser (Agilent Technologies), using RNA 6000 Nano Kit microchannel chips and reagents. cDNA synthesis (1-2 μg of RNA) and RT-PCR were performed using Moloney murine leukemia virus reverse transcriptase (200 U/μL; Promega) with random decamers and RETROscript kit (Ambion). Quantitative PCR was performed on the ABI Prism 7000 Sequence Detection System (Applied Biosystems). Reactions were set up in PCR MasterMix (Applied Biosystems), using specific primers for CDAN1/Cdan1 and reference GAPDH/Gapdh genes (Applied Biosystems). All reactions were performed in technical triplicate. Time-reaction data (ΔCt) were analyzed and normalized to the housekeeping genes GAPDH/Gapdh.
Generating a mouse model
The XG571 murine embryonic stem cell (mESC) clone was obtained from the International Gene-Trap consortium (www.genetrap.org) member BayGenomics at the University of California, San Diego, CA. This clone was independently confirmed to have a pGT1lxf vector inserted in the sequence of Cdan1 intron 25. This gene-trap vector encodes a splice-acceptor followed by β-galactosidase and neomycin-resistance gene sequences, and therefore constitutes a traceable tag for the murine codanin-1 expressed from the endogenous promoter. Confirmation of genotype and positional identification of the insertion of the β-geo gene-trap in Cdan1 were performed by multiplex PCR using unique oligonucleotide primers (supplemental Table 2) in optimized conditions.
The step-up technique of Mortensen et al19 was used in the attempt to obtain ES cells homozygous for the gene-trap. Embryoid bodies were differentiated as described by Wiles and Keller.20
Chimerae were generated by injection of the XG571 ES cells into CBA/C57BL6 blastocysts and transferred into pseudo-pregnant recipients. Male chimeras were bred to C57/BL6 mice, and agouti offspring were genotyped by multiplex PCR.
All animal work was carried out under Home Office United Kingdom license.
Results
Distribution and localization of codanin-1 shown by monoclonal antibody
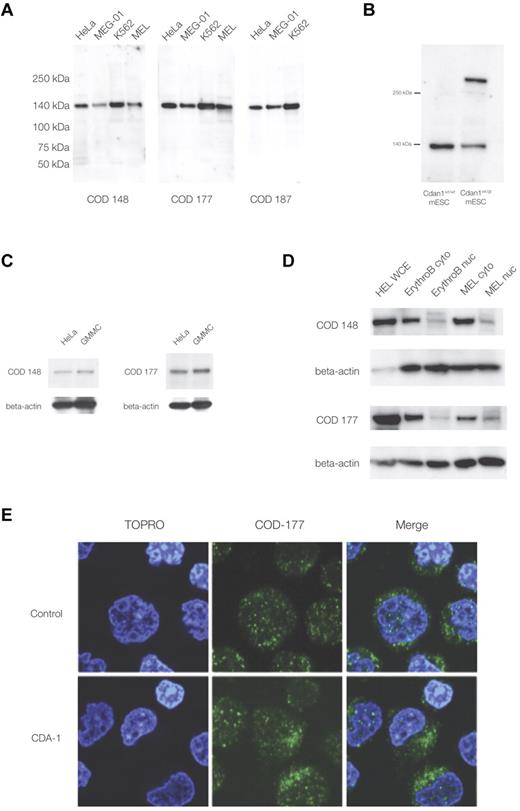
The study of codanin-1, and its distribution in normal and CDA-1 erythroblasts, has been hampered by the lack of a specific antibody. We have generated the first 3 monoclonal antibodies against codanin-1, each of a different IgG subclass. These antibodies all identified a single band of the appropriate size (∼ 134 kDa) in Western blots using protein extracts from human fibroblast, megakaryocytic, and erythroid cell lines (Figure 1A). Each antibody also identified a band of the same size in control mouse ES cells (E14, mESCs), and an additional band was seen in XG571 mESCs that had been modified by the insertion of a gene-trap vector into the Cdan1 gene (Figure 1B; see “A mouse model with a gene-trap plasmid in the Cdan1 gene” for details of the murine gene-trap of Cdan1). These results demonstrate the specificity of the antibodies for codanin-1 and its cross-reactivity across human and mouse cell extracts. In addition, when nonhematopoietic (ie, HeLa and GMMC) protein extracts were probed, the anti-codanin-1 antibodies were able to highlight the correct protein (Figure 1C). These findings show antibody specificity across species and confirm that codanin-1 is also expressed in nonerythroid tissue.
Western immunoblotting and immunofluorescence microscopy with murine monoclonal anti-codanin-1 (COD53, COD177, and COD187) antibodies. (A) Western (full-range) blot with the 3 uncloned hybridoma supernatants. Lanes correspond to HeLa, MEG-01, MEL, and K562 cell line protein extracts. (B) Western blot of whole-cell extracts of mESCs (E14) probed with the COD177 antibody. Bands of approximately 140 kDa (wild-type codanin-1) and more than 250 kDa (codanin-1/β-geo fusion) are visible. This demonstrates the specificity of the monoclonal antibody against the wild-type codanin-1 and a fusion protein of the expected size generated by gene-trapping one allele of Cdan1 (“A mouse model with a gene-trap plasmid in the Cdan1 gene”). (C) Western blot of nonerythroid brain cell line GMMC and HeLa cell line extracts. (D) Comparison of whole-cell extracts (WCE), cytoplasmic (cyto), and nuclear extracts (nuc) from cultured intermediate erythroblasts (ErythroB), and MEL cell line. (E) Immunofluorescence staining of control and CDA-1 primary human erythroblasts using the COD-177 antibody. DNA is counterstained with TOPRO-3.
Western immunoblotting and immunofluorescence microscopy with murine monoclonal anti-codanin-1 (COD53, COD177, and COD187) antibodies. (A) Western (full-range) blot with the 3 uncloned hybridoma supernatants. Lanes correspond to HeLa, MEG-01, MEL, and K562 cell line protein extracts. (B) Western blot of whole-cell extracts of mESCs (E14) probed with the COD177 antibody. Bands of approximately 140 kDa (wild-type codanin-1) and more than 250 kDa (codanin-1/β-geo fusion) are visible. This demonstrates the specificity of the monoclonal antibody against the wild-type codanin-1 and a fusion protein of the expected size generated by gene-trapping one allele of Cdan1 (“A mouse model with a gene-trap plasmid in the Cdan1 gene”). (C) Western blot of nonerythroid brain cell line GMMC and HeLa cell line extracts. (D) Comparison of whole-cell extracts (WCE), cytoplasmic (cyto), and nuclear extracts (nuc) from cultured intermediate erythroblasts (ErythroB), and MEL cell line. (E) Immunofluorescence staining of control and CDA-1 primary human erythroblasts using the COD-177 antibody. DNA is counterstained with TOPRO-3.
Normal erythroblast protein extracts showed the expected wild-type codanin-1 band on Western immunoblots. When separate nuclear and cytoplasmic extracts were prepared from these cells, more codanin-1 was found in the cytosol than in the nuclear fractions with all of the antibodies used (Figure 1D). This observation was confirmed in the murine erythroid MEL cell line. Immunofluorescence confirmed that, although codanin-1 is found in the nucleus, it is more abundant in the cytoplasm in primary human erythroblasts (Figure 1E). This localization pattern was unchanged in CDA-1 erythroblasts. Codanin-1 was found in similar quantities in primary erythroblasts of patients with CDA-1 and normal controls, as measured by Western blotting and immunofluorescence (supplemental Figure 1). Within the nucleus, codanin-1 is localized in foci, whereas the cytoplasmic protein sits in aggregations. Costaining with endoplasmic reticulum (ER) and Golgi apparatus markers indicated these organelles as probably locations (supplemental Figure 2). The localization of codanin-1 was found consistently with all 3 monoclonal antibodies. It is interesting to note the pattern of cytoplasmic staining to be slightly different in CDA-1 erythroblasts compared with normal controls. Although it is not possible to reliably quantify this observation, regions with concentrated foci of codanin-1 were observed in the cytoplasm of CDA-1 erythroblasts but not in the normal controls. Colocalization studies by immunofluorescence did not reveal whether codanin-1 might be abnormally concentrated in any of the cytosolic organelles.
Chromatin structure in primary CDA-1 erythroblasts
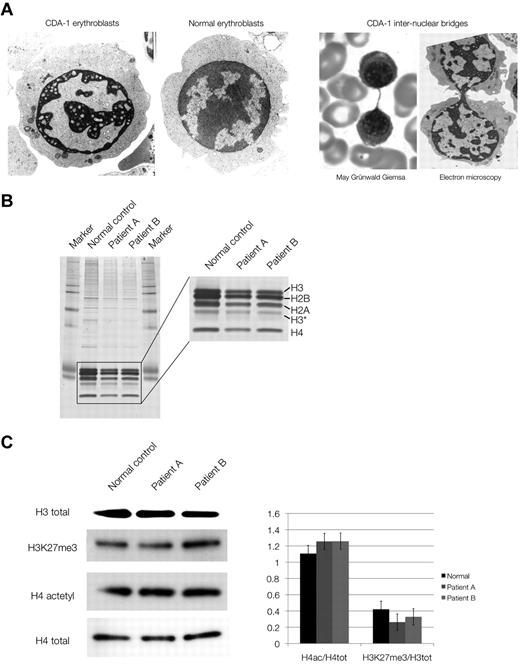
We studied the chromatin composition in human primary erythroblasts from 2 cases with CDA-1. The patients were Lebanese nonconsanguineous, homozygous carriers of the classic Bedouin mutation (3238C-T, R1040W) and have been described previously.3,21 Erythroblasts were cultured from patients' mononuclear cells by the 2-phase method of Fibach et al.10 Under these conditions, the intermediate erythroblasts recapitulate the CDA-1 bone marrow findings. Their chromatin has the characteristic spongy (“Swiss cheese”) appearance (Figure 2A) seen in this condition. The internuclear chromatin bridges can occasionally be observed, although not at the frequency seen in CDA-1 bone marrow aspirates, possibly because of the culture conditions.
Chromatin structure and composition in CDA-1 patients. (A) Electron microscopy of normal and CDA-1 cultured intermediate erythroblasts, and imaging of intranuclear chromatin bridging stained with May-Grünwald-Giemsa and by electron microscopy. (B) Silver staining of SDS-PAGE of histone extracts from cultured CDA-1 (patients A and B) and normal erythroblasts. (C) Western blots of total histones and histone marks. Quantification of histone marks relative to total histones (set to 1), adjusted to a normalized background.
Chromatin structure and composition in CDA-1 patients. (A) Electron microscopy of normal and CDA-1 cultured intermediate erythroblasts, and imaging of intranuclear chromatin bridging stained with May-Grünwald-Giemsa and by electron microscopy. (B) Silver staining of SDS-PAGE of histone extracts from cultured CDA-1 (patients A and B) and normal erythroblasts. (C) Western blots of total histones and histone marks. Quantification of histone marks relative to total histones (set to 1), adjusted to a normalized background.
Given the grossly abnormal chromatin structure in CDA-1 erythroblasts, we initially examined the overall composition of the chromatin by analyzing its histone component, which has not been previously described in CDA-1. No gross differences between normal and patient samples were seen (Figure 2B) or in the amounts of acetylated histones or various epigenetic marks of histone tails (H4 acetylation, H3K4 dimethylation [me2], H3K9 trimethylation [me3], and H3K27 trimethylation [me3]; Figure 2C) as tested by Western blotting and immunofluorescence. This suggests that histone signatures, involved in maintenance of chromatin and epigenetic regulation, are globally maintained in CDA-1. We confirmed the latter by ChIP-on-chip analysis where immunoprecipitated chromatin from CDA-1 erythroblasts and normal controls were hybridized to ENCODE genome-wide microrrays.22 Representative results of these experiments can be seen in supplemental Figures 3 and 4. A total of approximately 500 gene loci were examined in detail for the previously studied chromatin marks (H4 acetylation, H3K4 dimethylation [me2], H3K9 trimethylation [me3], and H3K27 trimethylation [me3]). No significant differences were seen, indicating that the epigenetic landscape is preserved in facultative heterochromatin of erythroblasts in CDA-1.
HP1α distribution in CDA-1
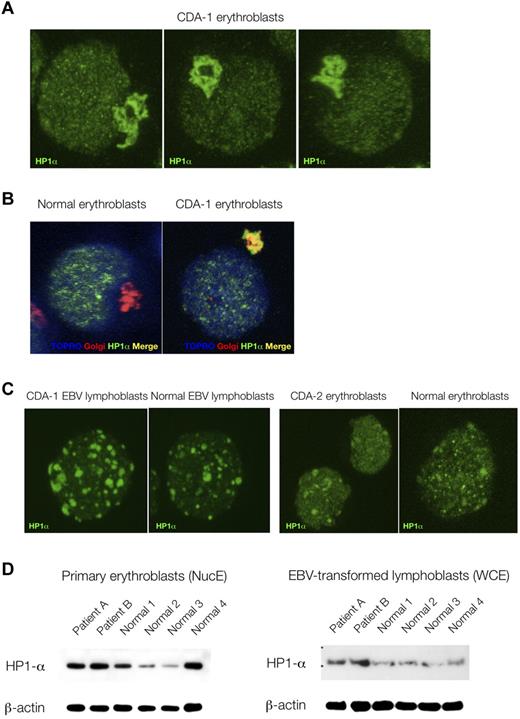
Because of the striking nuclear features displayed by CDA-1 patients' erythroblasts, we decided to compare the immunofluorescence localization patterns of various nuclear proteins, marking a variety of nuclear subcompartments. No differences were found between patients' and control erythroblasts in the distribution patterns of RNA-polymerase II CTD ser2P (H5), PML, nucleophosmin, HP1β, and HP1γ (data not shown). However, the localization of HP1α was found be markedly perturbed in the patients' samples. As can be seen in Figure 3A and B, HP1α accumulates and stains strongly in the Golgi apparatus of CDA-1 but not normal erythroblasts. This did not occur in EBV-derived lymphoblasts from CDA-1 patients, nor was it found in erythroblasts from a patient with CDA-2 (Figure 3C). We also were able to confirm that the abnormal localization of HP1α in CDA-1 patients is confined to the intermediate erythroblasts, where the spongy ultrastructural chromatin pattern is present (data not shown). Less mature (early/colony-forming unit-erythroid/proerythroblasts) and more mature erythroblasts (late/orthochromatic) did not show this pattern (data not shown). Furthermore, within the nucleus, HP1α foci display a microgranular pattern in CDA-1 erythroblasts (Figure 3A), whereas foci are larger and fewer in the normal controls (Figure 3C). We collected 3-dimensional confocal images and performed 2-dimensional profile measurements on the nuclei of CDA-1 and normal intermediate erythroblasts to confirm our subjective observation of these differences in the granular deposition pattern of HP1α (supplemental Figure 5). Although the qualitative distribution of HP1α is abnormal, the quantity of protein present in CDA-1 erythroblasts and EBV-derived lymphoblasts versus controls is at least maintained, if not increased (Figure 3D).
Abnormal accumulation of HP1α in CDA-1 erythroblasts is erythroid specific. (A) Accumulation of HP1α in CDA-1 intermediate erythroblasts by immunofluorescent microscopy. (B) Staining with Golgi-apparatus marker (giantin) confirms localization of HP1α in the Golgi apparatus. (C) Lack of abnormal localization in EBV-derived lymphoblasts from CDA-1 patients and CDA-2 intermediate erythroblasts. (D) Quantification of HP1α in primary erythroblasts and matched EBV-transformed lymphoblasts of CDA-1 patients and controls. Western blotting of nuclear extracts of primary erythroblasts (NucE; left) and of whole-cell extracts of EBV-transformed lymphoblasts (WCE; right).
Abnormal accumulation of HP1α in CDA-1 erythroblasts is erythroid specific. (A) Accumulation of HP1α in CDA-1 intermediate erythroblasts by immunofluorescent microscopy. (B) Staining with Golgi-apparatus marker (giantin) confirms localization of HP1α in the Golgi apparatus. (C) Lack of abnormal localization in EBV-derived lymphoblasts from CDA-1 patients and CDA-2 intermediate erythroblasts. (D) Quantification of HP1α in primary erythroblasts and matched EBV-transformed lymphoblasts of CDA-1 patients and controls. Western blotting of nuclear extracts of primary erythroblasts (NucE; left) and of whole-cell extracts of EBV-transformed lymphoblasts (WCE; right).
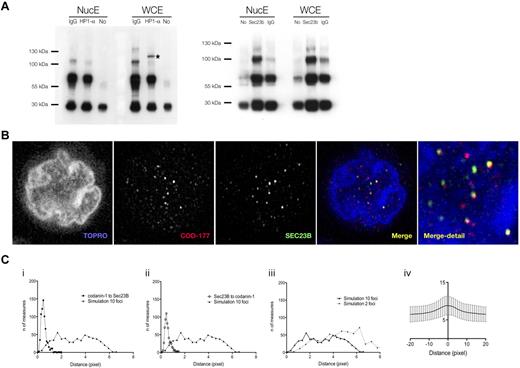
Our anti–codanin-1 antibodies were found to be unsuitable for immunoprecipitation. However, immunoprecipitation of erythroblast protein extracts with anti-HP1α antibodies coimmunoprecipitated codanin-1 from whole-cell extracts but not from nuclear extracts (Figure 4A). This interaction suggests that an abnormality in codanin-1 could be responsible for the aberrant localization of HP1α.
Colocalization of codanin-1 with HP1α and Sec23B. (A) Coimmunoprecipitation results. (Left) Whole-cell (WCE) and nuclear (NucE) protein extracts precipitated with the anti-HP1α antibody and blotted with the anti-codanin-1 antibody (COD177). (Right) Protein extracts (WCE, NucE) precipitated with the anti-Sec23B antibody and blotted with the anti-codanin-1 antibody (COD177). Nonspecific bands are immunoglobulins and their complexes. (B) Codanin-1 and Sec23B colocalization in an erythroblast by immunofluorescence. (Ci-ii) Nearest-neighbor distance measurements between codanin-1 and Sec23B foci (and reciprocally) extracted from confocal images from 600 cells. A histogram of simulated nearest-neighbor distance measurements generated in a random model of 10 foci/cell adjusted for volume and contour (cell and nucleus) is plotted in comparison. Each simulated histogram is generated by calculating distances from a random reference focus (Sec23B or codanin-1) to a nearest possible random neighbor focus (codanin-1 or Sec23B) in a virtual cell of similar cyotplasmic and nuclear proportions. The x-axis represents the distance in pixels; and y-axis, the number of distances measured. These graphs suggest that colocalization of codanin-1 and Sec23B is not random. (Ciii) The number of foci/cell impacts simulated histograms. Shown are 2 simulated histograms with either 2 or 10 foci/cell. In the 600 cells analyzed, the average number of foci was approximately 10. (Civ) Representation of Pearson colocalization model (X = 0 center of focus), suggesting colocalization of foci. Background noise accounts for low index values, and only trend is interpretable.
Colocalization of codanin-1 with HP1α and Sec23B. (A) Coimmunoprecipitation results. (Left) Whole-cell (WCE) and nuclear (NucE) protein extracts precipitated with the anti-HP1α antibody and blotted with the anti-codanin-1 antibody (COD177). (Right) Protein extracts (WCE, NucE) precipitated with the anti-Sec23B antibody and blotted with the anti-codanin-1 antibody (COD177). Nonspecific bands are immunoglobulins and their complexes. (B) Codanin-1 and Sec23B colocalization in an erythroblast by immunofluorescence. (Ci-ii) Nearest-neighbor distance measurements between codanin-1 and Sec23B foci (and reciprocally) extracted from confocal images from 600 cells. A histogram of simulated nearest-neighbor distance measurements generated in a random model of 10 foci/cell adjusted for volume and contour (cell and nucleus) is plotted in comparison. Each simulated histogram is generated by calculating distances from a random reference focus (Sec23B or codanin-1) to a nearest possible random neighbor focus (codanin-1 or Sec23B) in a virtual cell of similar cyotplasmic and nuclear proportions. The x-axis represents the distance in pixels; and y-axis, the number of distances measured. These graphs suggest that colocalization of codanin-1 and Sec23B is not random. (Ciii) The number of foci/cell impacts simulated histograms. Shown are 2 simulated histograms with either 2 or 10 foci/cell. In the 600 cells analyzed, the average number of foci was approximately 10. (Civ) Representation of Pearson colocalization model (X = 0 center of focus), suggesting colocalization of foci. Background noise accounts for low index values, and only trend is interpretable.
Codanin-1 and Sec23B colocalize
We found cytoplasmic codanin-1 to localize in a vesicle-like pattern, and in a cellular location suggestive of the ER. We therefore investigated potential links with the vesicular protein involved in the pathogenesis of CDA-2, Sec23B. Immunoprecipitation with anti-Sec23B antibodies did not clearly coprecipitate codanin-1, suggesting that they may not form a direct complex. However, by confocal immunofluorescence, there were clear foci within both the cytoplasm and nucleus of normal erythroblasts where the 2 colocalize (Figure 4B). Calculation of van Steensel's adaptation of the Pearson coefficient-based colocalization algorithm confirms a correlation between foci of Sec23B and codanin-1, and their comparison with a stochastic mathematical also excluded a random association (Figure 4C).23 Partial overlap of codanin-1 and Sec23B was also observed in CDA-1 and CDA-2 erythroblasts (data not shown).
A mouse model with a gene-trap plasmid in the Cdan1 gene
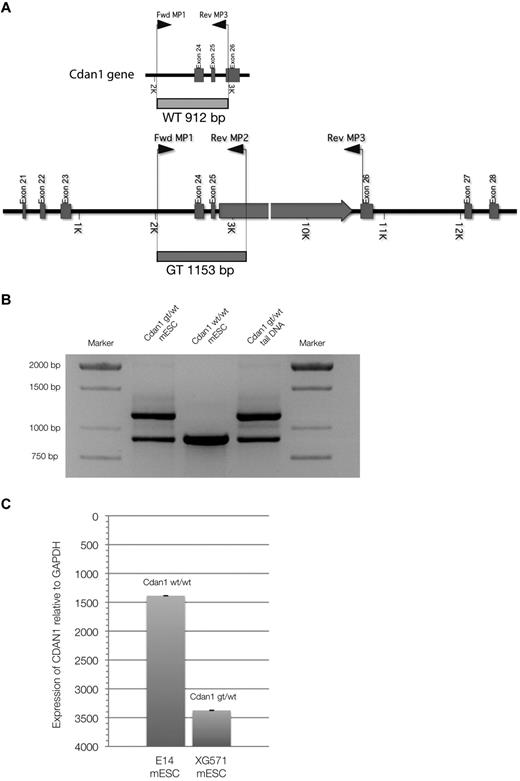
A mouse ES cell line containing a gene-trap plasmid in the Cdan1 gene, XG571-Cdan1gt/wt, was acquired from the International Gene-trap Consortium. This gene-trap inserts β-galactosidase and neomycin-resistance genes into the carboxy-terminal end of the codanin-1 protein, allowing tracking. We genotyped cells and mice using the multiplex PCR strategy as outlined in Figure 5A-B. By quantitative PCR expression analysis with specific Cdan1 primers and Western immunoblots using the novel monoclonal antibodies targeting codanin-1, we have shown that the expression of Cdan1 is reduced in the Cdan1gt/wtmESC cell line, but that codanin-1 and the fusion product of codanin-1 and the β-geo gene-trap are expressed (Figure 5C) and translated (Figure 1B). The fusion protein is detectable in Cdan1gt/wt mESCs by LacZ staining (data not shown).
Multiplex PCR genotyping strategy. (A) Schematic representation of primers and transcripts within the Cdan1 gene. (B) PCR products after multiplex PCR amplification. Amplification products from cell-line DNA of Cdan1gt/wt mESCs (positive control) and Cdan1wt/wt mESCs (E14, negative control) are compared with mouse tail-derived DNA to demonstrate stability of assay to identify murine live gene-trapped animals. (C) Codanin-1 mRNA expression of wild-type (E14, Cdan1wt/wt) and gene-trapped (XG571, Cdan1gt/wt) mESCs by quantitative PCR normalized to expression from housekeeping gene GAPDH.
Multiplex PCR genotyping strategy. (A) Schematic representation of primers and transcripts within the Cdan1 gene. (B) PCR products after multiplex PCR amplification. Amplification products from cell-line DNA of Cdan1gt/wt mESCs (positive control) and Cdan1wt/wt mESCs (E14, negative control) are compared with mouse tail-derived DNA to demonstrate stability of assay to identify murine live gene-trapped animals. (C) Codanin-1 mRNA expression of wild-type (E14, Cdan1wt/wt) and gene-trapped (XG571, Cdan1gt/wt) mESCs by quantitative PCR normalized to expression from housekeeping gene GAPDH.
Fewer embryoid bodies were generated from the Cdan1gt/wt mESCs compared with their parental E14 ES cell line, but hemoglobinization did not appear to be affected. However, no homozygous Cdan1gt/gt mESCs were obtained after stepping up the concentration of the neomycin selective agent (data not shown).
The unsuccessful attempts to obtain homozygous Cdan1gt/gt mESCs was mirrored by the lack of live-born Cdan1gt/gt mice. When pregnancies of heterozygous Cdan1wt/gt matings were terminated, no viable homozygous Cdan1gt/gt embryos were found (Table 1). Resorbed embryos in the proportion expected for homozygous Cdan1gt/gt were observed in pregnancies before 7.5 days of gestation. That death occurred before the onset of embryonic erythropoiesis (E7.5) implies a critical role of the protein during early development other than in erythropoiesis.
Results of gene-trapped Cdan1 mice breeding.
| . | Cdan1 genotype . | |||
|---|---|---|---|---|
| wt/wt, no. (%) . | gt/wt, no. (%) . | gt/gt, no. (%) . | Resorbed, no. (%) . | |
| Live offspring (n = 35) | 12 (34) | 23 (66) | 0 | NA |
| E12.5 (n = 50) | 10 (20) | 23 (46) | 0 | 17 (34) |
| E10.5 (n = 19) | 5 (26) | 9 (47) | 0 | 5 (27) |
| E9.5 (n = 9) | 1 (11) | 5 (55) | 0 | 3 (33) |
| E8.5 (n = 32) | 8 (25) | 18 (56) | 0 | 6 (19) |
| E7.5 (n = 8) | 2 (25) | 3 (37) | 1 (13) | 2 (25) |
| . | Cdan1 genotype . | |||
|---|---|---|---|---|
| wt/wt, no. (%) . | gt/wt, no. (%) . | gt/gt, no. (%) . | Resorbed, no. (%) . | |
| Live offspring (n = 35) | 12 (34) | 23 (66) | 0 | NA |
| E12.5 (n = 50) | 10 (20) | 23 (46) | 0 | 17 (34) |
| E10.5 (n = 19) | 5 (26) | 9 (47) | 0 | 5 (27) |
| E9.5 (n = 9) | 1 (11) | 5 (55) | 0 | 3 (33) |
| E8.5 (n = 32) | 8 (25) | 18 (56) | 0 | 6 (19) |
| E7.5 (n = 8) | 2 (25) | 3 (37) | 1 (13) | 2 (25) |
wt indicates wild-type; gt, gene-trap; and NA, not applicable.
In heterozygous Cdan1wt/gt embryos stained with X-Gal, Cdan1 is expressed ubiquitously during early embryogenesis (Figure 6A), with strong staining of the yolk sac hematopoietic blood islands (Figure 6B). High expression of Cdan1 was also observed in the cardiovascular and digestive systems of more mature embryos (Figure 6C; supplemental Figure 6).
X-Gal staining of whole heterozygote Cdan1gt/wt embryos. (A) Whole mounts at different stages of development. The only homozygous embryo Cdan1gt/gt, as shown in Table 1, obtained in this study clearly shows strong X-Gal staining, demonstrating Cdan1 expression from both gene-trapped alleles. (B) Intense X-Gal staining of yolk sac mesoderm-blood islands in Cdan1gt/wt embryos at E7.5 in cross section. (C) Intense X-Gal staining of a wide range of cell populations in Cdan1gt/wt embryos at E10.5. Bro indicates bronchus; Gu, gut; and YS, yolk sac.
X-Gal staining of whole heterozygote Cdan1gt/wt embryos. (A) Whole mounts at different stages of development. The only homozygous embryo Cdan1gt/gt, as shown in Table 1, obtained in this study clearly shows strong X-Gal staining, demonstrating Cdan1 expression from both gene-trapped alleles. (B) Intense X-Gal staining of yolk sac mesoderm-blood islands in Cdan1gt/wt embryos at E7.5 in cross section. (C) Intense X-Gal staining of a wide range of cell populations in Cdan1gt/wt embryos at E10.5. Bro indicates bronchus; Gu, gut; and YS, yolk sac.
Adult heterozygous Cdan1wt/gt mice had normal hematologic indices (supplemental Figure 7), red cell morphology (supplemental Figure 8), and did not display the characteristic ultrastructural findings of CDA-1 (data not shown). They have normal life expectancy and are fertile. No extrahematopoietic macroscopic phenotype could be identified. Analysis of the intracellular distribution of the codanin-1/β-geo fusion protein in erythroblasts of the heterozygous mice showed it to accumulate abnormally in the Golgi apparatus (supplemental Figure 9). This may be a result of the extra size of the fusion protein but may also indicate that codanin-1 normally transits through this organelle.
Discussion
CDAN1 is a highly conserved, widely expressed gene producing codanin-1, a protein lacking known protein motifs or domains, which might give clues to its function. Most cases of CDA-1, a hereditary anemia characterized by erythroblasts with abnormal chromatin structure, result from mutations in CDAN1.3
The ultrastructural phenotype in CDA-1 calls for an investigation of proteins involved in the higher-order structure of chromatin, and of potential consequences on gene regulation in mutants. Surprisingly, no studies of overall chromatin structure analyses in CDA patients have been reported in the literature.
In this study, experiments were performed on cultured erythroblasts of patients with CDA-1. These showed, for the first time, that the ultrastructural nuclear abnormalities were not associated with either alteration in histone composition or with changes in modifications of the epigenetic landscape over well-defined gene loci.
The major histone marks were remarkably well maintained in the analyzed regions, including H3K9me3, which has been shown to be crucial in HP1α binding to the nucleosome.24 One can therefore conclude that epigenetic regulation of the ENCODE contained loci is preserved in CDA-1. Nonetheless, it is important to underline that these genomic areas mainly constitute regions of euchromatin.
More interestingly, we have shown by immunofluorescence that the heterochromatin protein HP1α is sequestered in the Golgi apparatus in primary erythroblasts of patients with CDA-1. Moreover, the nuclear localization pattern of HP1α is abnormal in these cells. Interestingly, these phenomena appeared to be erythroid-specific (ie, not in CDA-1 EBV-derived lymphoblasts) and were not found in a patient with CDA-2. However, moderate nuclear differences cannot be excluded as this technique gives essentially qualitative analyses.
Furthermore, we were able to show the interaction of codanin-1 with HP1α by immunoprecipitation. In addition, we found that anti-HP1α antibodies coimmunoprecipitate codanin-1 from whole cell but not nuclear erythroblast extracts in CDA-1, and that reduced levels of codanin-1 can be measured in the nuclear fraction of such cells. Altogether, these observations could suggest that HP1α and codanin-1 might not be found in the same complex (or at the same proximity) depending on their nuclear or cytoplasmic location.
These novel findings suggest that codanin-1 mutations may cause CDA-1 via a secondary mechanism involving nuclear HP1α transport. Alternatively, codanin-1 mutations could lead to abnormal HP1α deposition on chromatin and subsequently cause a secondary inappropriate accumulation of HP1α in the Golgi apparatus. Further studies will be required to elucidate this. As a whole, our results suggest that HP1α plays a fundamental role in erythroid maturation. Perturbations of HP1α, a key player in heterochromatin structure and nuclear organization,25,26 could elegantly explain the ultrastructural phenotype observed in CDA-1 erythroblasts and these cells' difficulties with cytokinesis. Indeed, HP1α has been involved in a wide array of functions, mainly connecting the higher-order structure of chromatin and gene regulation.24,26 Interestingly, HP1α also seems to play a role in kinetochore formation and chromosome segregation.27 HP1α abnormalities could therefore account for the aberrant cytokinesis and the defect in chromatin structure in CDA-1 erythroblasts.
A previous study suggested that codanin-1 expression was restricted to the nucleus and varies with the cell cycle.28 Differences with the data presented by Noy-Lotan et al28 may have resulted from their use of a polyclonal rabbit antibody, whose specificity was not as clearly defined as for our series of murine monoclonal antibodies. Furthermore, these investigators' experiments were performed in nonerythroid cell lines, rather than in primary erythroid cells as studied here. Further work will be required to determine the significance of these differences.
If codanin-1 plays an essential role in transporting HP1α to the nucleus, how can the erythroid-specific phenotype of CDA-1 patients be explained? During terminal erythropoiesis, chromatin is progressively compacted, and previously facultative regions of the genome start to resemble constitutive heterochromatic regions. This is mediated by the propagation of heterochromatin proteins (of which HP1α is a key element) to euchromatic zones.29 A hypofunctional codanin-1 during terminal erythroid maturation could result in insufficient chromatin compaction, which ultimately would impair enucleation. The higher expression level of CDAN1 in erythroblasts, as shown by human and murine gene expression arrays, as well as our own murine and human quantitative PCR data (not shown in this study), may reflect the increased requirement for the protein in this tissue and hence the phenotypic effects. As a parallel of interest, it might be worth mentioning the observations in Diamond-Blackfan anemia. Mutations in the ribosomal protein genes, although ubiquitously expressed, can cause the erythroid-restricted phenotype of Diamond-Blackfan anemia.30,31 Altogether, these observations and ours could speculatively point to the highly specialized nature of erythroid maturation.
CDAs have been grouped because of the common dyserythropoiesis and similarities in the nuclear abnormalities of erythroblasts, but it is unclear whether they represent separate disorders or defects in different genes of a shared metabolic pathway. The preliminary evidence presented here, showing that codanin-1 partially colocalizes with Sec23B, the protein mutated in CDA-2, may suggest a possible molecular link between the 2 major types of CDA.4 Sec23B is a COP-II coatomer protein, a component of COP-II vesicles.32,33 These are involved in vesicular transport of proteins from the ER to the cis-Golgi, where proteins are modified post-translationally for secretion or integration into lysosomes, endosomes, or the cytoplasmic membrane.32 The observation that codanin-1 mutations affect the transport of HP1α through the Golgi apparatus suggests that there may be a link via intracellular transport pathways. Intriguingly, Sec23B and codanin-1 also colocalize in the nucleus. Previous studies have shown that Sec23B can be found in the nucleus, similar to other COP-II-associated proteins (ie, Sar1).33 One could speculate that Sec23B and codanin-1 are involved in an alternative, vesicle-independent, intranuclear pathway, although this needs to be confirmed by additional studies. Furthermore, abnormalities affecting the Golgi apparatus and/or the ER could compromise general protein transport. Stress of the ER/Golgi has recently been shown to enhance the secretion of hepcidin via CREBH, a liver-specific transcription factor.34 Therefore, ER/Golgi stress in hepatic cells could cause increased levels of hepcidin and be partly responsible for the iron overload observed in CDA-1.
Our mouse model, in which a gene-trap construct was inserted into the Cdan1 gene, generates a fusion product that can be detected at the RNA and protein levels in mESCs. This fusion protein should therefore contain normal codanin-1 sequence up to exon 25. Homozygosity for the gene-trap allele is embryonic lethal, either because the missing 3′ end is essential for function or because the large β-geo addition disrupts function sufficiently (including by abnormal protein folding) to effectively produce a null allele. This finding is consistent with the fact that homozygosity for a null CDAN1 allele has not been found in patients with CDA-1. Interestingly, as Cdan1gt/gt embryos die before the onset of primitive erythropoiesis (E7.5), codanin-1 must also have a critical role in developmental processes other than erythropoiesis. Indeed, within the gene-trapped murine Cdan1, the β-galactosidase insert provided a useful marker for examining expression of the gene in heterozygotes. We could confirm that Cdan1 is widely expressed in adult mice and that the highest levels can be measured in erythropoietic cells. During development, expression was also widespread with high levels of Cdan1 found in the digestive tract, the cardiovascular system, and parts of the neuroepithelium. At a cellular level, we observed that codanin-1 had an identical localization pattern in all examined tissues. Taken as a whole, we were surprised by these observations and think they deserve reporting. Indeed, although there seems to be no clinically observable gastrointestinal, cardiovascular, or neurologic phenotype in patients with CDA-1, we are not aware of any systematic studies in the literature. We think that a very comprehensive and careful workup of patients with CDA-1 might be required in light of these novel findings.
In conclusion, our results shed new light onto the function of codanin-1. Our studies show that the absence of this highly conserved ubiquitously expressed protein is embryonic lethal and that mutations responsible for the erythroid specific phenotype act via the abnormal cellular trafficking of the heterochromatin protein HP1α.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the Medical Research Council for continuing support. R.R. was supported by the Berrow Foundation & Oxford University, United Kingdom (Lord Florey Scholarship) and the Valeria Rossi di Montelera Foundation, Switzerland (Eugenio Litta Scholarship). D.J.P.F. is supported by an equipment grant from the Wellcome Trust. Sue Butler, Pik-Shan Li and Joanne Green provided core technical support in this study.
Authorship
Contribution: R.R. designed and performed the majority of experiments, analyzed and interpreted results, and wrote the paper; N.A.R. and M.D.G. performed and interpreted molecular experiments; J.A.S. supervised mouse production; L.E.B., J.C., and T.H. were responsible for antibody generation; J.S.-S. provided expertise in erythroblast culture; D.J.P.F. performed the electron microscopy experiments; J.M.B. and V.J.B. designed and supervised immunofluorescence experiments and cowrote the paper; D.R.H. designed experiments and edited the manuscript; and W.G.W. supervised experiments, analyzed and interpreted results, and cowrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Raffaele Renella, Jimmy Fund Clinic, Department of Pediatric Oncology, Dana 3, Dana-Farber Cancer Institute, 44 Binney St, Boston, MA 02115; e-mail: raffaele.renella@childrens.harvard.edu.