Abstract
Protein S has an important anticoagulant function by acting as a cofactor for activated protein C (APC). We recently reported that the EGF1 domain residue Asp95 is critical for APC cofactor function. In the present study, we examined whether additional interaction sites within the Gla domain of protein S might contribute to its APC cofactor function. We examined 4 residues, composing the previously reported “Face1” (N33S/P35T/E36A/Y39V) variant, as single point substitutions. Of these protein S variants, protein S E36A was found to be almost completely inactive using calibrated automated thrombography. In factor Va inactivation assays, protein S E36A had 89% reduced cofactor activity compared with wild-type protein S and was almost completely inactive in factor VIIIa inactivation; phospholipid binding was, however, normal. Glu36 lies outside the ω-loop that mediates Ca2+-dependent phospholipid binding. Using mass spectrometry, it was nevertheless confirmed that Glu36 is γ-carboxylated. Our finding that Gla36 is important for APC cofactor function, but not for phospholipid binding, defines a novel function (other than Ca2+ coordination/phospholipid binding) for a Gla residue in vitamin K–dependent proteins. It also suggests that residues within the Gla and EGF1 domains of protein S act cooperatively for its APC cofactor function.
Introduction
Protein S is an anticoagulant plasma protein recognized as a member of the protein C anticoagulant pathway, in which it acts as a cofactor for activated protein C (APC).1 Protein S mediates its APC cofactor activity by enhancing the inactivation of activated coagulation factors V (FVa) and VIII (FVIIIa). Protein S also inhibits thrombin generation independently of APC, which has recently been attributed to an enhancement of the activity of tissue factor pathway inhibitor (TFPI).2,3 Numerous studies have shown an association between protein S deficiency and an increased risk for thrombosis.4-9 The importance of protein S in vivo has been confirmed in mouse models where mice die in utero of coagulopathy and hemorrhages after knocking out the gene-encoding protein S (PROS1).10,11
Protein S is a 635-amino acid vitamin K–dependent glycoprotein. It circulates in plasma at a concentration of 350nM, with approximately 60% being in complex with C4b-binding protein.12 It has generally been accepted that only free protein S can mediate cofactor function for APC,13,14 but this has recently been questioned by Maurissen et al who showed that the protein S–C4b-binding protein complex impairs APC-catalyzed proteolysis of FVa at Arg506, whereas it enhances the APC-catalyzed proteolysis at Arg306.15
Protein S is divided into a number of structural domains composing an N-terminal vitamin K–dependent Gla domain, a thrombin-sensitive region (TSR), 4 epidermal growth factor (EGF)–like domains, and a region homologous to sex hormone binding globulin domain.1,13 In vitamin K–dependent proteins, γ-carboxylation of glutamic acid residues leading to formation of Gla residues occurs during biosynthesis.16 These residues are important for calcium binding and proper folding of the Gla domain.16 The correctly folded Gla domain binds to negatively charged phospholipid membranes, which is essential for the function of protein S. The N-terminal Gla domain in protein S contains 11 potential Gla sites (at amino acids 6, 7, 14, 16, 19, 20, 25, 26, 29, 32, and 36).16 Grinnell et al showed that a reduction of as few as 2 of these Gla residues led to a dramatic decrease in APC cofactor function and binding to phospholipids.17 Glu36, although assumed to be a Gla residue,18 is not conserved in other vitamin K–dependent plasma proteins and is not predicted to coordinate calcium.16,19
Despite the importance of protein S as an anticoagulant protein, the exact molecular mechanisms of its APC cofactor function are still unknown. APC cleaves FVa at Arg306, Arg506, and Arg679.20 It has been shown that protein S accelerates APC inactivation of FVa mainly through enhancement of the cleavage at Arg306.21,22 Yegneswaran et al have suggested that binding of protein S to APC leads to a relocation of the active site of APC, enabling more efficient cleavage of FVa in position 306.23 Lately, additional APC cleavage sites have been found in FVa. Protein S was equally efficient as a cofactor for proteolysis at each of these novel sites, arguing against the relocation hypothesis.24 Several studies identifying important regions of protein S for the cofactor function toward APC have been published. Domain-specific monoclonal antibodies,25 deletions of whole domains,26,27 and amino acid residue substitutions28 have all suggested an interaction site for APC somewhere in the N-terminal domains of protein S (Gla-TSR-EGF1-EGF2). Saller et al created 2 composite protein S variants (termed Face1 and Face2) by replacing 4 and 7 amino acids, respectively, with the corresponding residues in the prothrombin Gla domain.29 Both variants lacked APC cofactor function. However, protein S Face1 was reported to not bind to phospholipid membranes, and it was therefore not possible to conclude whether these residues were important for cofactor function toward APC.29 In a recent report, we identified a critical region in the EGF1 domain, involving Asp95 together with Asp78 and Gln79, as being essential for APC cofactor function.30
In the present study, we extend our recently published data on Asp9530 by drawing on the work done by Saller et al29 to examine the importance of solvent exposed amino acid residues in the Gla domain for the APC cofactor function of protein S. We identified Gla36 as important for protein S APC cofactor function but not being required for normal phospholipid binding. These findings therefore support a cooperative model, with Gla36 in the Gla domain, and Asp95 in the EGF1 domain independently being essential for the APC cofactor function of protein S.
Methods
Mutagenesis of protein S
The cDNA encoding wild-type (WT) protein S had previously been introduced into a pcDNA6 vector (Invitrogen).31 Protein S mutants were constructed through site directed mutagenesis (KOD Hot Start DNA polymerase kit; Novagen) using the pcDNA6/protein S vector as a template and mutagenic oligonucleotide primers (Thermo Scientific). All mutations were verified by DNA sequencing. The following individual and composite point mutations were introduced: N33S, P35T, E36A, Y39V, Face1 (a variant with all these mutations) and E36D.
Protein S expression
The pcDNA6/protein S vector containing either the cDNA for WT protein S or mutated variants was either transiently or stably transfected into HEK293T or HEK293 cells (ATCC), respectively. Stably transfected cells were selected with 5 μg/mL Blasticidin-HCl (Invitrogen). Protein S variants were expressed in OptiMEM I (Invitrogen) supplemented with 10 μg/mL vitamin K (Roche Products) as previously described.30 Media containing protein S was harvested, dialyzed in 20mM Tris-HCl pH 7.5, 0.15M NaCl (TBS), 3mM CaCl2 and concentrated, as required.
Purification of protein S variants
Protein S and its variants were either partially purified by barium citrate precipitation32 or extensively purified using anion-exchange chromatography as an additional purification step. For this, the partially purified protein was applied on a HiTrap DEAE fast-flow column (5 mL; GE Healthcare) equilibrated with TBS. After washing with the same buffer, protein S was eluted with a 40 mL of linear CaCl2 gradient (0-30mM). Fractions were analyzed using 10% SDS-PAGE under nonreducing conditions, and those containing pure protein S were pooled and dialyzed in TBS, 3mM CaCl2. Both the partially and extensively purified protein S samples were concentrated, as required, and the protein S concentration determined using a protein S ELISA.
Protein S ELISA
An in-house sandwich ELISA for protein S was used to quantify protein S. A 96-well Maxisorp plate (Nunc) was coated with 1 μg/mL polyclonal rabbit anti–protein S antibody (Dako) in 50mM sodium carbonate buffer, pH 9.6, at 4°C overnight. Washing steps were performed in triplicate with 250 μL of TBS, 3mM CaCl2, 0.1% Tween 20 between each step. The plates were quenched with 2% BSA (Sigma-Aldrich) in TBS, 3mM CaCl2 for 2 hours at 37°C. A plasma standard curve was generated to provide 0 to 2nM protein S dilutions to quantify protein S variants in media, whereas plasma purified protein S (Enzyme Research Laboratories) was used to quantify partially and extensively purified protein S variants.
All samples were diluted in TBS, 3mM CaCl2, 0.5% BSA, and incubated for 1 hour at 37°C. Bound protein S was detected by a mouse monoclonal anti–protein S antibody (4nM, Haematologic Technologies Inc), followed by a horseradish peroxidase-conjugated goat anti–mouse antibody (8nM, Dako). The plate was developed with o-phenylenediamine dihyrochloride (Sigma-Aldrich), according to the manufacturer's instruction. The enzymatic reaction was terminated with H2SO4 and the absorbance read at 492 nm. The standard curve was fitted using nonlinear regression for a sigmoidal dose-response model using GraphPad Prism Version 4.02.
Phospholipid vesicle preparation
Phospholipid vesicles were prepared from phospholipids (Avanti Polar Lipids) dissolved in chloroform by extrusion essentially as described previously.30,33 Synthetic phospholipids 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-dioleoyl-sn-glycero-3-phosphoserine (DOPS), and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) were used in the plasma assay, phospholipid binding assay, and in the FXa inhibition assay. Natural phospholipids L-α-phosphatidylserine (brain extract), L-α-phosphatidylethanolamine (egg extract), and L-α-phosphatidylcholine (egg extract) were used in the FVa inactivation assay, the prothrombinase assay, the FVIIIa inactivation assay, and the Xase assay.
Binding of protein S phospholipids
Phospholipid binding assays were performed to determine the apparent dissociation constants (Kd app) for interactions between protein S and phospholipid membranes, as previously described.30 In brief, protein S variants (0-120nM), partially or extensively purified, were incubated in plates coated with phospholipid vesicles (DOPS/DOPC/DOPE, 20:60:20), and bound protein S was detected using a polyclonal peroxidase-conjugated anti–protein S antibody (Affinity Biologicals). All incubation steps were performed in the presence of 3mM CaCl2, and the wells were washed between each step. To confirm a specific interaction, the assay was also performed in the presence of 10mM ethylenediaminetetraacetic acid (EDTA) and in wells without phospholipids. Phospholipid binding was also studied, using surface plasmon resonance. Liposomes were immobilized to a L1 sensor chip surfaces (GE Healthcare) using a Biacore T100 (Biacore) to a response of 5000 to 7000 response units, as previously described.34 The presence of a procoagulant binding surface was confirmed by factor Xa association and dissociation, which both were predictably monoexponential.
Binding of protein S to domain-specific monoclonal antibodies
Kd app for interactions between protein S and domain-specific monoclonal antibodies were determined as previously described.30 Briefly, the monoclonal domain-specific anti–protein S antibodies MK21 (Gla domain), MK47 (Gla domain), or MK67 (TSR domain) were coated in a 96-well plate.25 Increasing concentrations of protein S was added to the wells, and bound protein was detected in 2 steps by a rabbit polyclonal antibody against protein S (Dako) followed by a goat anti–rabbit horseradish peroxidase-conjugated antibody (Dako). Washing steps were performed between each step, and all incubation steps were performed in the presence of 3mM CaCl2.
Thrombin generation assay by CAT
The APC cofactor function of the protein S variants was assessed by calibrated automated thrombography (CAT) using a Fluoroskan Ascent FL plate reader (Thermo Labsystem) in combination with Thrombinoscope software (Synapse BV) as described previously.30 Thrombin generation was initiated in protein S–depleted plasma (Affinity Biologicals) by 1pM tissue factor (Dade Innovin, Dade Behring), 50μM phospholipid vesicles (DOPS/DOPC/DOPE, 20:60:20), and 16.6mM CaCl2. Thrombin generation was monitored using 0.42mM of the fluorogenic substrate Z-GlyArg-AMC-HCl (Bachem) in the presence or absence of APC (Xigris) and protein S (9 and 0-120nM, respectively). To inhibit contact activation, 65 μg/mL plasma corn trypsin inhibitor was added. A polyclonal antibody directed against TFPI (100nM, Haematologic Technologies Inc) was added to inhibit TFPI during the assay.
Protein S–dependent APC-mediated FVa inactivation assay
Protein S enhancement of APC-mediated inactivation of the FV variant, FV R506Q/R679Q, was assessed by determining the loss of FVa cofactor activity. The experiments were performed essentially as described previously.30 Briefly, FV was activated by human thrombin, followed by the addition of hirudin. The activated FVa variant was used as a substrate for APC to determine the ability of protein S to enhance the cleavage at Arg306 in FVa by APC in the presence of phospholipids. The inactivation reaction was stopped by a dilution in ice-cold HN BSA Ca2+ (5mM CaCl2, 5 mg/mL BSA, 25mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid. pH 7.7, 0.15M NaCl [HN]). The remaining FVa activity was determined using a prothrombinase assay.30 An aliquot of the FVa inactivation reaction was incubated with phospholipid vesicles, FXa, and prothrombin in the presence of CaCl2. The amount of thrombin generated was measured kinetically by the cleavage of the chromogenic substrate S-2238 (Chromogenix) at 405 nm for 15 minutes.
To derive the rate constant for FVa cleavage, time course experiments were performed. FVa (0.8nM) was incubated with 25μM phospholipids in the presence and absence of 0.25nM APC and 100nM protein S. For calculation of the pseudo–first-order rate constant for APC-mediated cleavage at Arg306, equation 1 was used as previously described33 :
Vat = Va0 × −k306 × t + C × Va0 × (1−e−k306 × t)
Vat and Va0 represent the cofactor activity of FVa determined at time t and at time point 0, respectively. C is the remaining activity of FVa after cleavage at position 306, and k306 is the rate constant for cleavage at position 306.
Protein S–dependent APC-mediated FVIIIa inactivation assay
Enhanced APC-mediated inactivation of FVIIIa by purified protein S was followed by determining the loss of FVIIIa cofactor activity toward FIXa in the conversion of FX to FXa. A FVIIIa inactivation assay, initially described by Shen and Dahlbäck35 and modified by Evenäs et al,36 was used here with minor modifications. FVIII (0.7 U/mL) was activated by incubation in a reaction mix (9nM FIXa and 88μM phospholipid vesicles, L-X-phosphatidylserine/L-X-phosphatidylcholine/L-X-phosphatidylethanolamine in a ratio of 10:70:20, in HN BSA Ca2+) with human thrombin (0.1 U/mL) for 3 minutes at 37°C. The reaction was stopped by the addition of hirudin (0.29 U/mL), and FVIIIa was diluted 1:1.4 in the reaction mix. Cleavage of FVIIIa by APC was determined by mixing 60 μL of the reaction mix containing FVIIIa with 45 μL of APC, FV, and increasing concentrations of protein S. Final concentrations were 1, 2.5, and 0 to 100nM, respectively. The final concentrations of FVIIIa, FIXa, and phospholipids in the inactivation step were 280 mU/mL, 5nM, and 50μM, respectively. The inactivation reaction was allowed to proceed for 2.5 minutes at 37°C, and then 20 μL of bovine FX was added for an Xase assay (final concentration of 0.5μM). The remaining FVIIIa was allowed to activate FX to FXa for 3 minutes at 37°C until the reaction was stopped by the addition of 125 μL of EDTA buffer (50mM Tris, 100mM NaCl, 20mM EDTA, 1% PEG 6000, pH 7.9). The amount of FXa generated was measured kinetically by the cleavage of the chromogenic substrate S-2765 (Chromogenix) at 405 nm for 15 minutes.
Mass spectrometry to assess γ-carboxylation of protein S Glu36
A 20-μg aliquot of purified protein S was dissolved in 50 μL of 50mM ammonium hydrogen carbonate buffer (Ambic buffer, pH 7.8). Freeze-dried sequencing grade modified trypsin (Promega) was dissolved in 20 μL of the Ambic buffer and added into the sample at a 1/50 (weight:weight) protease/protein ratio. The sample was incubated for 10 hours at 37°C. Disulphide bond reduction was carried out by addition of 10 μL of 1 mg/mL dithiothreitol in Ambic buffer with incubation for 60 minutes at 37°C. Free cysteine alkylation was achieved by addition of 10 μL of 30 mg/mL iodoacetic acid in the Ambic buffer with incubation for 60 minutes in the dark at room temperature. The protein S tryptic digest was acidified to pH 6.0 by addition of 0.1% trifluoroacetic acid (Romil) before liquid chromatography-mass spectrometry (LC-MS) analyses.
On-line LC-MS analyses were performed using a reverse-phase C18 nano-LC connected to an ElectroSpray Ionisation Quadrupole–Time of Flight (ESI Q/TOF) mass spectrometer (API ESI Q-STAR Pulsar; MDS Sciex). Tryptic digests of protein S were loaded and separated on an analytical PepMap C18 nanocapillary column (15 cm length, 75 mm internal diameter) fitted on an Ultimate HPLC chain (LC Packings). The flow rate was 0.15 μL/minute. After the sample loading onto the column, sequential elution of the peptides was carried out using solvent A (0.1% [volume:volume] trifluoroacetic acid in 2% [volume:volume] acetonitrile) and solvent B (0.1% [volume:volume] trifluoroacetic acid in 90% [volume:volume] acetonitrile), with the following gradient: 0 to 5 minutes, 99% A; 5 to 6 minutes, 99% to 90% A; 6 to 25 minutes, 90% to 60% A; 25 to 30 minutes, 60% to 50% A; 30 to 60 minutes, 50% to 25% A; 60 to 75 minutes, 25% to 5% A; 75 to 85 minutes, 5% A; 85 to 86 minutes, 5% to 95% A; and 86 to 90 minutes, 95% A. Data were acquired using the Analyst QS software Version 1.1 with an automatic information-dependent acquisition function.
Protein S–dependent TFPI-mediated FXa inhibition assay
The TFPI cofactor activities of protein S and protein S E36A were assessed using a FXa inhibition assay. CaCl2 (5mM), 10μM phospholipid vesicles (DOPS/DOPC/DOPE, 20:60:20), and 200μM S-2765 were mixed with or without protein S (0-80nM) and purified TFPI37 (1nM; kind gift from Dr Tsutomu Hamuro, the Chemo-Sero-Therapeutic Research Institute, Kaketsuken, Japan). The reaction was started on addition of 0.5nM FXa, and the FXa activity was measured kinetically by the cleavage of the chromogenic substrate S-2765 (Chromogenix) at 405 nm for 30 minutes. All dilutions were made in TBS and 5 mg/mL BSA, and all reported concentrations are final.
Results
Evaluation of APC cofactor function of protein S variants substituted in the Gla domain by CAT
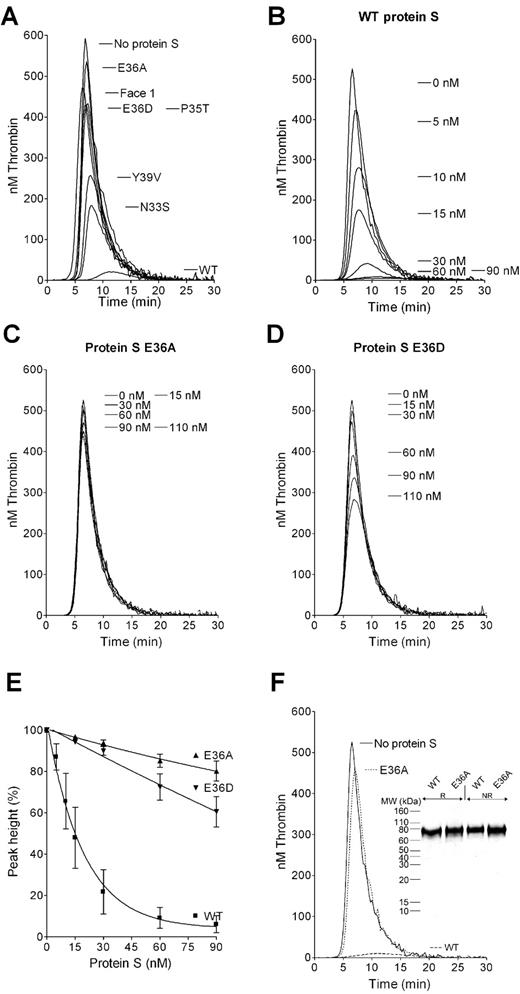
Thrombin generation using CAT was used to study APC cofactor function of protein S variants. Protein S–deficient plasma was supplemented with recombinant protein S from concentrated and dialyzed conditioned media samples. The screening of WT protein S and its variants for their APC cofactor function was performed at 9nM APC and 60nM protein S. Under these conditions, WT protein S almost completely inhibited thrombin generation (Figure 1A). The composite variant, protein S Face1, had very limited cofactor function for APC confirming results obtained using different methodology.29 Protein S Face1 is a composite variant containing 4 amino acid substitutions. The importance for APC cofactor function of each of the 4 amino acid residues was studied using single residue substitutions of N33S, P35T, E36A, and Y39V. The protein S N33S and Y39V variants had modestly impaired APC cofactor function, whereas protein S E36A and P35T variants were almost completely inactive (Figure 1A). As Glu36 is predicted to be a Gla residue, another variant was created, E36D. Aspartic acid is similar to glutamic acid but cannot be γ-carboxylated. Protein S E36D also showed limited inhibitory effect on thrombin formation (Figure 1A).
Effect of protein S variants for APC cofactor activity on thrombin generation. Thrombin generation was measured in protein S–deficient plasma supplemented with 9nM APC and 60nM of protein S variants (A), 0 to 90nM WT protein S (B), 0 to 110nM protein S E36A (C), or 0 to 110nM protein S E36D (D). Typical experiments are shown (n = 3). Dose-response data from titrations with WT protein S, protein S E36A, and protein S E36D (0-90nM) in the presence of 9nM APC is shown in panel E. Data are mean ± SEM; n = 4. (F) Thrombin generation in protein S free plasma supplemented with 9nM APC and 60nM purified protein S. (Inset) Purified WT protein S and protein S E36A on a silver-stained 10% SDS-PAGE under reducing (R) and nonreducing (NR) conditions.
Effect of protein S variants for APC cofactor activity on thrombin generation. Thrombin generation was measured in protein S–deficient plasma supplemented with 9nM APC and 60nM of protein S variants (A), 0 to 90nM WT protein S (B), 0 to 110nM protein S E36A (C), or 0 to 110nM protein S E36D (D). Typical experiments are shown (n = 3). Dose-response data from titrations with WT protein S, protein S E36A, and protein S E36D (0-90nM) in the presence of 9nM APC is shown in panel E. Data are mean ± SEM; n = 4. (F) Thrombin generation in protein S free plasma supplemented with 9nM APC and 60nM purified protein S. (Inset) Purified WT protein S and protein S E36A on a silver-stained 10% SDS-PAGE under reducing (R) and nonreducing (NR) conditions.
Titrations (0-110nM) of protein S E36A and E36D were performed in parallel with WT protein S (Figure 1B-D), using 9nM APC. Protein S E36A had negligible APC cofactor function even at concentrations as high as 110nM (Figure 1C). The E36D substitution in protein S did not completely abolish but severely reduced its cofactor function (Figure 1D). Whereas WT protein S reduced the peak height by approximately 80% at a concentration of 30nM, the same concentration of protein S E36D only reduced it by 10% (Figure 1E). All experiments presented in Figure 1A through E were conducted using protein S in concentrated conditioned media. These results were confirmed using purified proteins. WT protein S and protein S E36A variants were purified by barium citrate precipitation, followed by anion exchange chromatography. The proteins migrated as single bands under reducing and nonreducing conditions on SDS-PAGE and were judged to be approximately 95% pure (Figure 1F inset). As in concentrated culture media, 60nM WT protein S, in the presence of 9nM APC, almost completely inhibited thrombin generation. Addition of 60nM protein S E36A to APC had a minimal effect, replicating the findings with the protein S in conditioned media (Figure 1F). Under these experimental conditions, protein S had no effect on thrombin generation in the absence of APC (data not shown).
Phospholipid binding of WT protein S, protein S Face1, and protein S E36A
Binding to negatively charged phospholipid surfaces is crucial for protein S to mediate its APC cofactor function. Protein S Face1 has previously been shown to bind phospholipid vesicles with 9 to 10 times lower affinity than WT protein S.29 We used a solid-phase assay to evaluate the ability of the protein S variants to bind phospholipid vesicles. WT protein S bound efficiently to DOPS/DOPC/DOPE vesicles with Kd app of 3.80 ± 1.03 and 3.77 ± 1.93nM for barium citrate precipitated and highly purified protein, respectively. As expected, binding was calcium dependent, and no binding occurred in the presence of EDTA or in the absence of phospholipids (data not shown). Surprisingly, both protein S Face1 and E36A bound phospholipid vesicles equally well as WT protein S with Kd app of 3.92 ± 1.03 and 3.80 ± 1.68nM, respectively (Table 1; P > .05). Binding of protein S E36A to phospholipid surfaces was confirmed using surface plasmon resonance. The complex, biphasic nature of adsorption to the phospholipids bilayer made the binding affinity impossible to quantify (data not shown). Taken together, the solid-phase assay and the surface plasmon resonance showed that the lack of APC cofactor function of both protein S Face1 and E36A did not arise from decreased binding to phospholipids.
Binding of protein S to phospholipid vesicles and domain-specific monoclonal antibodies
| . | WT . | Face1 . | 36A . |
|---|---|---|---|
| Phospholipid vesicles | 3.80 ± 1.03 | 3.92 ± 1.03 | 3.80 ± 1.68 |
| MK21 (Gla) | 3.11 ± 0.51 | 8.54 ± 0.62 | 4.88 ± 0.19 |
| MK47 (Gla) | 2.23 ± 0.57 | 4.48 ± 0.93 | 2.80 ± 0.71 |
| MK67 (TSR) | 0.45 ± 0.03 | 0.75 ± 0.03 | 0.50 ± 0.07 |
| . | WT . | Face1 . | 36A . |
|---|---|---|---|
| Phospholipid vesicles | 3.80 ± 1.03 | 3.92 ± 1.03 | 3.80 ± 1.68 |
| MK21 (Gla) | 3.11 ± 0.51 | 8.54 ± 0.62 | 4.88 ± 0.19 |
| MK47 (Gla) | 2.23 ± 0.57 | 4.48 ± 0.93 | 2.80 ± 0.71 |
| MK67 (TSR) | 0.45 ± 0.03 | 0.75 ± 0.03 | 0.50 ± 0.07 |
Kdapp values are given in nanomoles (mean ± SD) from 3 independent experiments.
Binding to domain-specific antibodies
We compared the affinities of protein S Face1 and protein S E36A to that of WT protein S for immobilized domain-specific antibodies in a solid-phase assay. Protein S in concentrated conditioned media was evaluated for binding to antibodies directed against either the Gla domain (MK21 and MK47) or the TSR domain (MK67). Kd app values were obtained by fitting the binding curves to a 1-site binding equation. There was no significant difference (P > .05) between any of the protein S variants for any of the 3 antibodies tested (Table 1) using the Mann-Whitney test, suggesting that the residue substitutions did not lead to any major conformational change in the Gla and TSR domains of the protein.
Protein S enhancement of APC-mediated cleavage of FVa at Arg306
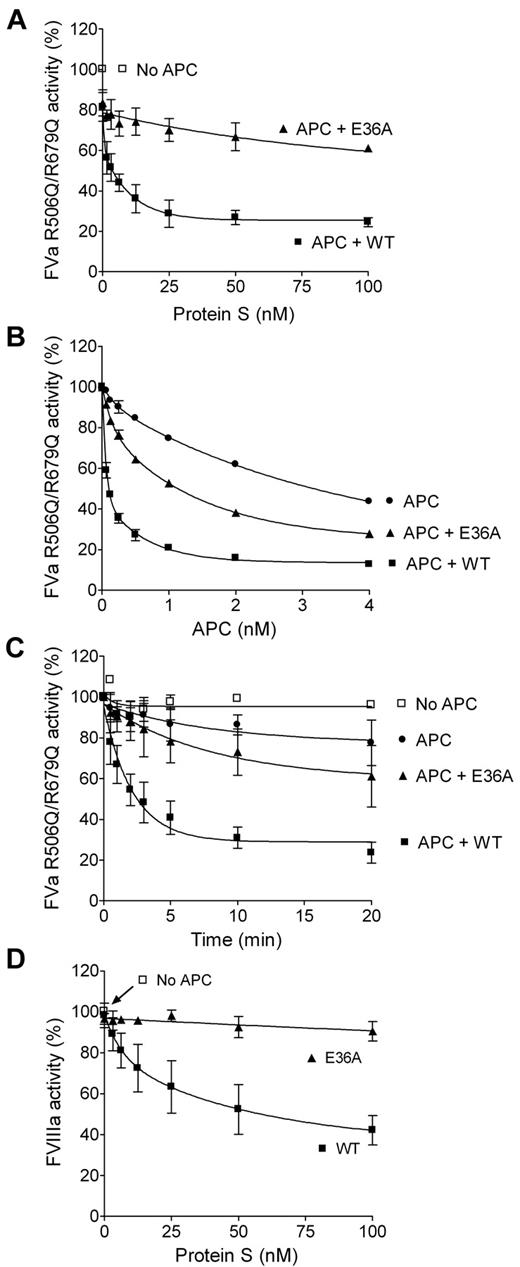
To evaluate the ability of the protein S variants to enhance APC-mediated cleavage of FVa at Arg306, a FVa inactivation assay with the R506Q/R679Q FVa variant was used. The FVa inactivation was performed in the presence and absence of 0.25nM APC with 0 to 120nM protein S. The remaining FVa activity after a 10-minute inactivation was measured in a prothrombinase assay. WT protein S efficiently enhanced the APC-mediated cleavage of Arg306 in FVa, in contrast to protein S E36A (Figure 2A). After inactivation using 25nM WT protein S, only 27% of the FVa activity remained, whereas as much as 60% FVa activity remained after inactivation with 100nM protein S E36A. When titrating APC (0-4nM) and keeping the protein S concentration constant (100nM), it was shown that 4nM of APC and 100nM protein S E36A was needed to decrease the FVa activity to the same levels as obtained from 0.25nM APC and 100nM WT protein S (Figure 2B). Protein S had no effect on the FVa activity in the absence of APC (data not shown).
Protein S enhancement of APC-mediated cleavage of FVa at Arg306 and of FVIIIa. Protein S was incubated with 0.8nM FVa R506Q/R679Q and phospholipids in the presence or absence of APC. The remaining FVa activity was measured in a prothrombinase assay (A-C). The FVa activity was measured either after incubation with increasing concentrations of protein S (0-100nM) in the presence or absence of 0.25nM APC (n = 3; A) or after incubation with 0 to 4nM APC in the presence or absence of 100nM protein S (n = 2; B). Time course experiments were performed to calculate the pseudo–first-order rate constants of WT protein S and protein S E36A using 100nM protein S and 0.25nM APC (Table 2; n = 4; C). Protein S (0-100nM) was incubated with 280 mU/mL FVIIIa, 5nM FIXa, 2.5nM FV, and phospholipids in the presence or absence of 1nM APC for 2.5 minutes. The remaining FVIIIa activity was measured with an Xase assay (n = 3; D). Data are plotted as mean ± SD.
Protein S enhancement of APC-mediated cleavage of FVa at Arg306 and of FVIIIa. Protein S was incubated with 0.8nM FVa R506Q/R679Q and phospholipids in the presence or absence of APC. The remaining FVa activity was measured in a prothrombinase assay (A-C). The FVa activity was measured either after incubation with increasing concentrations of protein S (0-100nM) in the presence or absence of 0.25nM APC (n = 3; A) or after incubation with 0 to 4nM APC in the presence or absence of 100nM protein S (n = 2; B). Time course experiments were performed to calculate the pseudo–first-order rate constants of WT protein S and protein S E36A using 100nM protein S and 0.25nM APC (Table 2; n = 4; C). Protein S (0-100nM) was incubated with 280 mU/mL FVIIIa, 5nM FIXa, 2.5nM FV, and phospholipids in the presence or absence of 1nM APC for 2.5 minutes. The remaining FVIIIa activity was measured with an Xase assay (n = 3; D). Data are plotted as mean ± SD.
To quantify the difference in rate of inactivation of FVa through cleavage at Arg306 in the presence and absence of WT protein S and protein S E36A, time course experiments were performed (Figure 2C). FVa activity was measured at different time points (0-20 minutes) after incubation in the presence or absence of 0.25nM APC and in the presence or absence of 100nM protein S. The remaining FVa activity was measured using the prothrombinase assay. The FVa activity at different time points was used to calculate the pseudo–first-order rate constant for the inactivation of FVa R506Q/R679Q, and these were then corrected for the APC concentration used (Figure 2C). APC-mediated cleavage at FVa Arg306 was enhanced 24.7-fold by WT protein S but only 2.75-fold by protein S E36A (Table 2). The pseudo–first-order constants were also derived for 4nM APC in the presence and absence of protein S E36A. These data were not significantly different from the ones derived when using 0.25nM APC (data not shown).
Pseudo-first-order rate constants (k) for the APC-catalyzed inactivation of FVa R506Q/R679Q in the presence and absence of WT protein S and protein S E36A
| . | No protein S . | WT protein S . | Protein S E36A . |
|---|---|---|---|
| k (M−1̇ s−1) | 1.63 ± 0.92 × 106 | 3.77 ± 1.66 × 107 | 4.15 ± 3.18 × 106 |
| Fold enhancement | 1 | 24.7 ± 5.34 | 2.75 ± 1.76 |
| . | No protein S . | WT protein S . | Protein S E36A . |
|---|---|---|---|
| k (M−1̇ s−1) | 1.63 ± 0.92 × 106 | 3.77 ± 1.66 × 107 | 4.15 ± 3.18 × 106 |
| Fold enhancement | 1 | 24.7 ± 5.34 | 2.75 ± 1.76 |
Rate constants are given as mean ± SD from 4 independent experiments.
Protein S enhancement of APC-mediated cleavage of FVIIIa
A FVIIIa inactivation assay was used to study the ability of the protein S variants to enhance APC-mediated inactivation of FVIIIa. The remaining FVIIIa activity, after 2.5 minutes inactivation with 0 to 100nM protein S, 2.5nM FV in the presence or absence of 1nM APC, was measured in an Xase assay. WT protein S clearly enhanced the APC-mediated cleavage of FVIIIa, whereas protein S E36A did not seem to have any effect under the conditions used (Figure 2D). Only 43% of the FVIIIa activity remained after inactivation using 100nM WT protein S, whereas more than 90% activity remained after inactivation with protein S E36A, using the same conditions. Protein S had no effect on the FVIIIa activity in the absence of APC (data not shown).
Evaluation of the γ-carboxylation of Glu36 determined by mass spectrometry
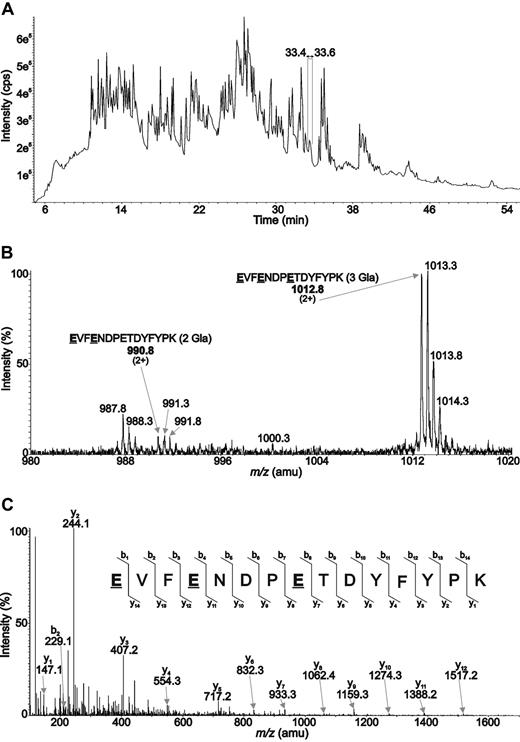
As Glu36 is not conserved among vitamin K–dependent proteins, we used mass spectrometry to determine whether or not Glu36 in the protein S Gla domain is γ-carboxylated. Pure WT protein S was slowly digested by trypsin, followed by nano-LC ESI/MS and subsequent sequencing by MS/MS analyses. Two doubly charged [M+2H]2+ molecular ions were detected at m/z 990.8 and 1012.8, respectively (t = 33.4 minutes, Figure 3A), indicating the presence of 2 species of the 29Glu-Lys43 tryptic peptide. In these fragments, either 2 (m/z 990.8) or 3 (m/z 1012.8) of 3 glutamic acid residues were modified by carboxylation. The amino acid sequence in these 2 fragments was confirmed by a similar fragmentation pattern of the 2 ions. Ion abundancy showed that Gla36 was predominantly γ-carboxylated because all 3 glutamic acid residues are carboxylated in 80% of the 29Glu-Lys43 tryptic peptides.
Characterization of the Gla36 residue within the protein S amino acid sequence by mass spectrometry. Protein S was incubated with trypsin, and the subsequent tryptic digest was analyzed by LC-MS/MS using an analytical C18 nanocapillary column plugged to an electrospray quadrupole–time of flight mass spectrometer (ESI Q-STAR). Two distinct species of the EVFENDPETDYFYPK peptide, containing 2 and 3 γ-carboxylated glutamic acid residues, were detected between 33.4 and 33.6 minutes elution time as shown in the TIC chromatogram (A). These 2 peptides were observed in positive ion mode as doubly charged molecular ions with m/z 990.76 and 1012.75, respectively. (One example of the 3 possible combinations of the 2 Gla containing peptide at m/z 990.8 is shown; B.) Both of these molecular ions were subjected to collision-induced fragmentation (parent ion m/z 1012.75 in panel C). The resulting MS/MS spectra show a number of fragment ions, mainly b- and y-type ions, allowing the unambiguous characterization of the peptide amino acid sequence.
Characterization of the Gla36 residue within the protein S amino acid sequence by mass spectrometry. Protein S was incubated with trypsin, and the subsequent tryptic digest was analyzed by LC-MS/MS using an analytical C18 nanocapillary column plugged to an electrospray quadrupole–time of flight mass spectrometer (ESI Q-STAR). Two distinct species of the EVFENDPETDYFYPK peptide, containing 2 and 3 γ-carboxylated glutamic acid residues, were detected between 33.4 and 33.6 minutes elution time as shown in the TIC chromatogram (A). These 2 peptides were observed in positive ion mode as doubly charged molecular ions with m/z 990.76 and 1012.75, respectively. (One example of the 3 possible combinations of the 2 Gla containing peptide at m/z 990.8 is shown; B.) Both of these molecular ions were subjected to collision-induced fragmentation (parent ion m/z 1012.75 in panel C). The resulting MS/MS spectra show a number of fragment ions, mainly b- and y-type ions, allowing the unambiguous characterization of the peptide amino acid sequence.
Evaluation of TFPI cofactor activity of protein S E36A in a purified FXa inhibition assay
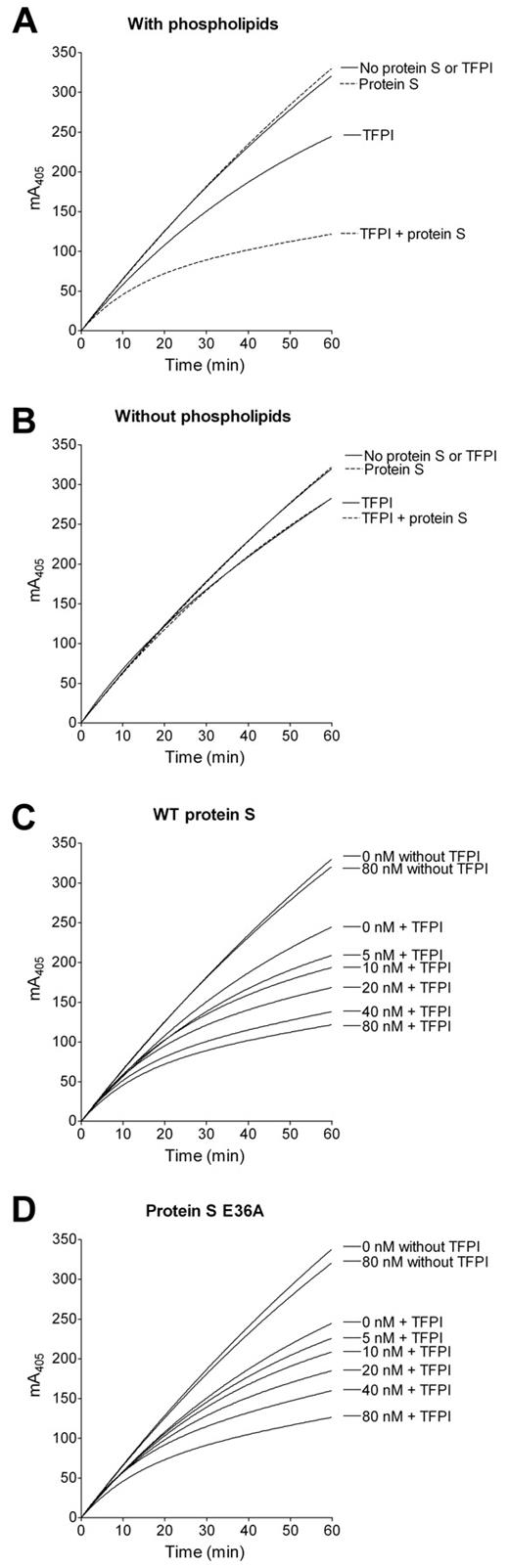
The importance of Gla36 for TFPI cofactor activity of protein S was assessed through the inhibition of FXa activity. As expected, protein S enhanced TFPI in a phospholipid-dependent manner (Figure 4A-B).2 Protein S had no effect on FXa activity in the absence of TFPI (Figure 4). When adding increasing amounts of protein S (0-80nM) in the presence of a fixed concentration of TFPI (1nM) and FXa (0.5nM), both WT protein S and protein S E36A efficiently enhanced TFPI-mediated inactivation of FXa (Figure 4C-D).
Effect of WT protein S and protein S E36A on TFPI cofactor activity through inhibition of FXa. Conversion of 200μM S-2765 by 0.5nM FXa was monitored by the absorbance at 405 nm in the presence (A) or absence (B) of 10μM phospholipids, 1nM TFPI, and 80nM WT protein S. The reaction was started on addition of FXa. WT protein S (C) and protein S E36A (D) were titrated (0-80nM) to the reaction containing 1nM TFPI (n = 3). Representative experiments are shown.
Effect of WT protein S and protein S E36A on TFPI cofactor activity through inhibition of FXa. Conversion of 200μM S-2765 by 0.5nM FXa was monitored by the absorbance at 405 nm in the presence (A) or absence (B) of 10μM phospholipids, 1nM TFPI, and 80nM WT protein S. The reaction was started on addition of FXa. WT protein S (C) and protein S E36A (D) were titrated (0-80nM) to the reaction containing 1nM TFPI (n = 3). Representative experiments are shown.
Discussion
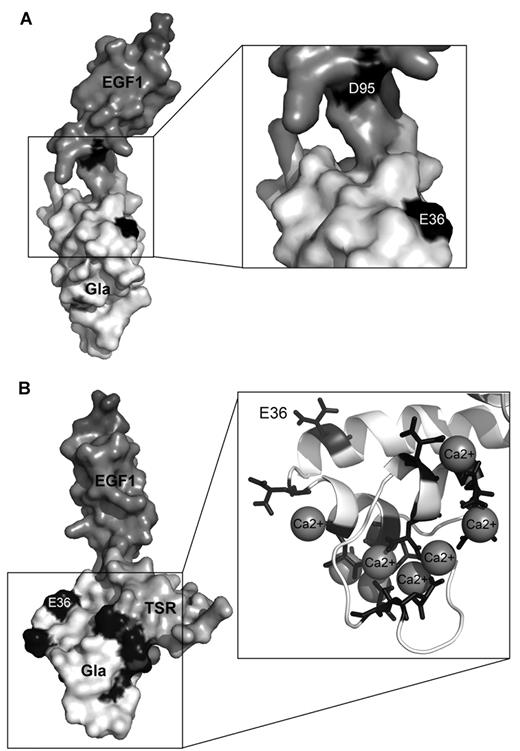
That protein S plays an essential role in the regulation of coagulation is well established. It acts as a cofactor to APC by enhancing its inactivation of FVa and also FVIIIa. Despite its clinical and physiologic relevance, the molecular basis of the protein S cofactor function is not completely defined. We have previously shown that protein S D95A has greatly reduced cofactor function for APC in plasma and for APC-mediated cleavage at Arg306 in FVa. We proposed that Asp95 within EGF1 of protein S can function as an interaction site for APC.30 Protein S Face1, a composite variant where 4 amino acid residues within the Gla domain of protein S have been substituted with the corresponding residues in prothrombin, has previously been shown to have decreased anticoagulant function in a plasma clotting assay.29 We have confirmed the decreased APC cofactor function using CAT. However, in contrast to the results of Saller et al,29 we find a normal binding of protein S Face1 to phospholipid membranes. Because of this, the variant is potentially more interesting than what was previously thought. Interestingly, the amino acid residues substituted in protein S Face1 are located on the same side of protein S as Asp95 in a molecular model of the Gla-TSR-EGF1 domains (Figure 5A). When studying the 4 amino acid changes of protein S Face1, as single point substitutions, we found that the lack of anticoagulant function of protein S Face1 was mainly because of 2 of the 4 residue substitutions, P35T and E36A. As prolines are important for protein folding/stability, it is difficult to draw any firm conclusions concerning the P35T substitution. Glu36, like all glutamic acid residues in the Gla domain (Figure 5B), is generally assumed to be γ-carboxylated.16 In this study, we confirmed the γ-carboxylation of Glu36 in our recombinant WT protein S by use of advanced mass spectrometry techniques (Figure 3). Furthermore, protein S E36A bound to phospholipid surfaces normally suggesting correct folding of the Gla domain (Table 1). This was shown in a solid-phase assay and further confirmed by the normal activity of protein S E36A in the phospholipid-dependent FXa inhibition assay (Figure 4). Protein S E36D, nevertheless, had severely reduced APC cofactor function compared with WT protein S.
Location of Gla36 within the protein S Gla-TSR-EGF1 model in respect to Asp95 and other Gla residues. Domains are labeled with different shades of gray. Gla36 (depicted as E36; in the Gla domain) and Asp95 (D95; in the EGF1 domain) are highlighted in black (A). All 11 Gla residues are in black. The boxes show the location of the Gla residues and predicted coordinated calcium ions in more detail in a cartoon model (B). The model is taken from Villoutreix et al.19
Location of Gla36 within the protein S Gla-TSR-EGF1 model in respect to Asp95 and other Gla residues. Domains are labeled with different shades of gray. Gla36 (depicted as E36; in the Gla domain) and Asp95 (D95; in the EGF1 domain) are highlighted in black (A). All 11 Gla residues are in black. The boxes show the location of the Gla residues and predicted coordinated calcium ions in more detail in a cartoon model (B). The model is taken from Villoutreix et al.19
Protein S is known to multimerize during purification, which can influence its phospholipid-dependent interactions in vitro.38,39 Accordingly, the plasma-based assays were performed using both protein S in conditioned media and extensively purified protein S. No difference in APC cofactor function was observed between protein S in conditioned media or fully purified protein S. Moreover, protein S E36A displayed minimal APC cofactor function, regardless of its purification status.
Protein S mediates its APC cofactor activity by enhancing its inactivation of both FVa and FVIIIa.35,40 Protein S accelerates FVa inactivation by mainly promoting the slow cleavage at Arg306, whereas the Arg506 cleavage is only weakly potentiated by protein S.21,22 In a FVa inactivation assay using a FVa variant, FVa R506Q/R679Q, which allowed us to selectively analyze cleavage of FVa in position Arg306, we show that WT protein S efficiently enhances APC activity, whereas protein S E36A had minimal effect (Figure 2). Using time course assays, we estimated that WT protein S enhances APC-mediated cleavage of FVa Arg306 by 24.7-fold, compared with 2.75-fold for protein S E36A (Table 2). Accordingly, protein S E36A only has 11% of the activity of WT protein S, which is similar to that previously reported for protein S 95A (13%).30 The effect of protein S E36A on APC-mediated inactivation of FVIIIa was investigated using a FVIIIa inactivation assay. A dose-dependent effect on FVIIIa inactivation was seen for WT protein S, whereas no enhancing activity was seen for protein S E36A, suggesting a similarly important role for Gla36 in APC-mediated inactivation of both FVa and FVIIIa.
Protein S is also a cofactor for TFPI, by enhancement of its inhibition of FXa.2,3 To study the TFPI cofactor function of protein S E36A, we performed a FXa inhibition assay. In these assays, protein S E36A behaved similarly to WT protein S. As the ability of protein S to enhance FXa inhibition by TFPI in this assay is strictly dependent on phospholipids, this confirms that the E36A substitution does not affect phospholipid binding or the overall conformation of the protein. Most importantly, this also suggests that different amino acids on protein S mediate cofactor function for either APC or TFPI, respectively.
Gla residues are traditionally associated with blood coagulation through the binding of vitamin K–dependent proteins to negatively charged phospholipid surfaces.16 However, Gla residues have also been identified in peptides from invertebrates,41,42 which are thought to have evolved before blood coagulation. Most hypothesized roles of Gla residues in the many proteins involve coordination of calcium.41,43,44 In contrast, Gla36 in protein S is not predicted to coordinate calcium19 (Figure 5B), neither is it involved in phospholipid binding of protein S. In a model of Gla-TSR-EGF1 domains of protein S,19 Gla36 lies outside of the ω-loop and is predicted to point away from the phospholipid membrane (Figure 5B). Collectively, this suggests that the γ-carboxylation of Glu36 is essential for protein S function and is involved in the interactions through which protein S enhances APC function. Taken together with the previously reported results for Asp95 in APC cofactor function, this study suggests an extended interaction surface on protein S spanning its Gla and EGF1 domains (Figure 5A) that mediates cofactor function of APC.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Ms Sofia Carlsson for valuable discussions.
This work was supported by the British Heart Foundation (PG/09/105), Novartis (unrestricted educational grant), and the Swedish Research Council (grant 07143).
National Institutes of Health
Authorship
Contribution: J.A., H.M.A., K.C., E.N., B.D., M.P., H.R.M., J.T.B.C. and D.A.L. designed the research, analyzed the results, and wrote the paper; and J.A., H.M.A., K.C., E.N., and Y.Y. performed the experiments.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Josefin Ahnström, Imperial College London, Centre for Haematology 5th Floor, Commonwealth Bldg, Hammersmith Hospital Campus, Du Cane Rd, London, W12 0NN, United Kingdom; e-mail: j.ahnstrom@imperial.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal