Abstract
Faithful repair of DNA lesions is a crucial task that dividing cells must actively perform to maintain genome integrity. Strikingly, nucleotide excision repair (NER), the most versatile DNA repair system, is specifically down-regulated in terminally differentiated cells. This prompted us to examine whether NER attenuation might be a common feature of all G0-arrested cells, and in particular of those that retain the capacity to reenter cell cycle and might thus convert unrepaired DNA lesions into mutations, a prerequisite for malignant transformation. Here we report that quiescent primary human B lymphocytes down-regulate NER at the global genome level while maintaining proficient repair of constitutively expressed genes. Quiescent B cells exposed to an environment that causes both DNA damage and proliferation accumulate point mutations in silent and inducible genes crucial for cell replication and differentiation, such as BCL6 and Cyclin D2. Similar to differentiated cells, NER attenuation in quiescent cells is associated with incomplete phosphorylation of the ubiquitin activating enzyme Ube1, which is required for proficient NER. Our data establish a mechanistic link between NER attenuation during quiescence and cell mutagenesis and also support the concept that oncogenic events targeting cell cycle- or activation-induced genes might initiate genomic instability and lymphomagenesis.
Introduction
B-cell malignancies result from multiple oncogenic events, including chromosomal translocations, deletions, amplifications, and point mutations in genes crucial for B lymphocyte development and differentiation. Several B-cell–specific mechanisms are thought to contribute to the 2 main steps in carcinogenesis: mutagenic events and subsequent proliferation. Initiation of B-cell transformation and further proliferation of malignant B cells occur after antigenic stimulation through the B-cell receptor, which is normally involved in B-cell–mediated immune responses against bacterial and viral infections.1,2 In addition, germinal center B cells actively down-regulate p53 to prevent the genetic stress caused by rearranging Ig genes during cell proliferation from triggering apoptosis. This rescue mechanism also renders them unusually tolerant to DNA damage in general.3 On the other hand, somatic hypermutation and class switch recombination, both required for the generation of high-affinity functional antibodies by germinal center B cells, may also be responsible for aberrant mutations in non-Ig genes and for chromosomal translocation between the Ig loci and a proto-oncogene, two common features of transformed B cells.2
Although there is increasingly compelling evidence for the latter mechanism in the pathogenesis of lymphoma, it is limited to defined regions in particular genes and is unlikely to constitute the sole source of mutations in B cells.4,5 Like any other cell, B lymphocytes are subject to various types of DNA damage that can lead to an accumulation of mutations when not repaired. Among the many DNA repair systems, nucleotide excision repair (NER) is probably the most versatile and can handle a vast array of DNA lesions of endogenous or environmental origin.6 The paramount importance of NER for genomic stability is illustrated by the high incidence of cancer in xeroderma pigmentosum (XP), a recessive disease resulting from mutations in NER genes.7 Yet, despite its importance, NER is known to be down-regulated in several cell types, eg, neurons, smooth and striated muscle, adipocytes, and keratinocytes.8 We have recently elucidated the mechanism of this down-regulation: a lack in ubiquitination of an NER enzyme, most likely a component of the TFIIH complex, resulting from a decrease in phosphorylation of the ubiquitin-activating enzyme Ube1.9
A common characteristic of the aforementioned cells is that they are terminally differentiated and never need to replicate their DNA. One could thus reason that such cells may dispense with the burden of repairing the bulk of their genome, as long as they maintain the integrity of the genes they are using. We have previously shown that it is indeed the case, and that differentiated cells concentrate their remaining NER activity inside transcription factories,10,11 thereby insuring proficient repair of active genes. This phenomenon, which we have termed transcription domain-associated repair (DAR), is genetically distinct from transcription-coupled repair (TCR) and likely exists to ensure proficient repair of both strands of active genes in an otherwise repair-deficient background.11,12 By contrast, TCR uses RNA polymerase II as a lesion sensor and thus provides for faster repair of the transcribed strand only.6
We wondered whether the aforementioned repair phenotype might be common to all G0-arrested cells. Contrarily to terminally differentiated cells, naive B lymphocytes are nondividing cells that retain the ability to proliferate, should they receive the proper stimulus. One would thus expect NER to remain proficient in these cells, even during their quiescent phase, as the accumulation of unrepaired DNA damage would likely translate in a high rate of mutagenesis when these cells resume proliferation. Yet, we report here that quiescent human B lymphocytes display a marked NER deficiency, which likely proceeds from the same mechanism as in differentiated cells. We also demonstrate that stimulation of B lymphocytes after DNA damage results in a high rate of mutagenesis, similar to that observed in XP cells. We postulate that NER attenuation during quiescence might contribute to the pathogenesis of B-cell malignancies.
Methods
Cell culture and UV irradiation
Primary human B lymphocytes were isolated from buffy coats, which were obtained from the United Kingdom National Blood Service. Each buffy coat was a pool from 2 donors. The total number of donors per experiment is shown in the figure legends. PBMCs were separated by differential migration on Ficoll-Paque Plus (GE Healthcare) and depleted from monocytes by adherence to culture dishes. The lymphocyte population was incubated with human CD43 Microbeads and passed through LS columns on a MACS Separator (Miltenyi Biotec Inc). Quiescent B cells, which are CD43-negative, were thereby collected “untouched” and cultured in RPMI medium (Lonza) supplemented with 10% FCS (SCI), 100mM penicillin, 100mM streptomycin, 100mM glutamine, 1mM sodium pyruvate, 100μM nonessential amino acids, and 5.5 × 10−5M 2-mercaptoethanol (all supplements from Invitrogen). Quiescent B cells (2 × 106 cells/mL) were induced to proliferate with 10 μg/mL affiniPure goat anti-human IgA + IgG + IgM (H + L) F(ab′)2 fragments (Jackson) and 500 ng/mL Pam3CSK4 bacterial product (InvivoGen). Unless otherwise stated, proliferating cells were used 72 hours later. GM01646 (XP-E; Coriell) and GM1953 (wild-type; Coriell) B lymphoblasts were cultured in RPMI with 10% FCS, 100mM penicillin, 100mM streptomycin, and 100mM glutamine.
For UV irradiation, quiescent or proliferating B lymphocytes, as well as GM01646 (XP-E) and GM01953 (WT) B lymphoblasts, were resuspended in PBS (Invitrogen) and irradiated for the appropriate time with a 254-nm germicidal lamp at a fluency of 7-8 μW/cm2. The Ube1-specific inhibitor Pyr-41 (Calbiochem) was dissolved in DMSO and added to the culture medium 2 hours before irradiation.
To measure cell proliferation, cells were exposed (or not) to 5 J/m2 UV in PBS, resuspended in culture medium, and dispensed into a 96-well plate at 2 × 106 cells/mL for primary B lymphocytes and 2 × 105 cells/mL for lymphoblasts. At various times, cells were labeled for 20 hours with 1 μCi [3H]thymidine and harvested with a Filtermate 196 cell harvester (Packard) onto DNA-binding GF/C filters (Whatman). The filters were layered with Microscint PS fluid (PerkinElmer) and counted in a Microbeta scintillation counter (Packard).
Flow cytometry
Flow cytometry analyses were performed with FACSCalibur or FACSAria. Purity of primary B cells was assessed by double staining with mouse anti-human IgD FITC (IgG2a κ; BD Biosciences) and mouse anti-human CD19 PE (IgG1 κ; BD Biosciences). For propidium iodide staining, cells were fixed overnight at 4°C in 70% ethanol, then washed with PBS (Invitrogen) and incubated in 50 μg/mL propidium iodide (Sigma-Aldrich) and 50 Unit/mL RNase A (Sigma-Aldrich) for 15 minutes at room temperature. For differential staining of DNA and RNA, cells were fixed as described previously, then rinsed in HBSS containing Mg2+ and Ca2+ (Invitrogen), and incubated with 2 μg/mL Hoechst 33342 (Sigma-Aldrich) and 4 μg/mL Pyronin Y (Sigma-Aldrich) for 20 minutes at a density of 2 × 106 cell/mL.
To appraise cellular division, 24 hours after irradiation (or not) cells were labeled with 1μM CFSE (Sigma-Aldrich) in serum-containing culture medium, incubated at 37°C for 10 minutes, and quenched by adding an equal volume of FCS and incubating at room temperature for 10 minutes. Cells were washed 3 times with medium and then cultured for the indicated times before flow cytometry and analysis with FlowJo software, excluding dead cells on the basis of propidium iodide staining.
NER assays
Mutagenesis assay
UV-induced mutations were detected by a modification of the method described by Kaur et al.14 DNA regions of interest were amplified from 12 μg of genomic DNA with high-fidelity DNA polymerase HF-2 Advantage (Clontech) under the following conditions: 94°C for 30 seconds, 10 cycles of 94°C for 20 seconds, 65°C for 20 seconds, and 68°C for 20 seconds with the annealing temperature decreasing 1°C per cycle (touch down); 15 cycles of 94°C for 10 seconds, 55°C for 20 seconds, and 68°C for 20 seconds; 68°C for 6 minutes; 4°C hold. The forward (F) and reverse (R) primers used for amplification of genomic fragments were as follows: Myosin (NCBI number M57965.2), F: CCCAAACCCTAACTTTTCTTTCTC, R: CCATGGAGATAGTTGGTCTCAGTC; DHFR (NG_023304.1), F: AGCACCCCAGATGAAAGAATAGTA, R: AAGTCTTGCATGATCCTTGTCAC; TP53 (X54156), F: AGTACTCCCCTGCCCTCAA, R: CCACTCGGATAAGATGCTGAG; Cyclin D2 (AF518005), F: TGGACGCGTCTCTCTCTTTC R: ACGCGTCTCTCTCTTTC; and BCL6 (NG_007149), F: CCGAAGATTAGTCCCACGTC, R: CTCTCCTCCACCTCCTTTCC.
Amplified fragments were purified from 1.8% agarose gels with a Qiaquick gel extraction kit (QIAGEN). Purified PCR products (500 ng) were digested with 1 Unit of Tsp509I (New England Biolabs) for 1 hour at 65°C, and purified with Qiaquick PCR purification kit (QIAGEN). Digested products were incubated with linkers AACTGTGCTATCCGAGGGAATGGAACCTGAGTCCTCTCACCGCA (4 μL from a 4 mg/mL stock) and AATTTGCGGTGA) (4 μL from a 1 mg/mL stock), and 5 μL of T4 DNA ligase buffer (10×) in a final volume of 49 μL. The mix was incubated at 50°C for 5 minutes, slowly cooled to 10°C (1°C/min), and incubated at 16°C for 30 minutes with 1-3 μL of T4 DNA ligase (2000 Unit/μL; New England Biolabs). Ligated products were amplified by nested PCR with Titanium Taq polymerase (Clontech), first with primer specific for the 20 first bases of the linker (AACTGTGCTATCCGAGGGAA) and one of the primers (forward or reverse) previously used for amplification of genomic fragments, then 5 μL of the first reaction was amplified again with a nested primer specific for both the linker and the restriction site (CTGAGTCCTCTCACCGCAAATT) and a primer specific for the genomic fragment. Cycling conditions: 94°C for 30 seconds, 25 cycles of 94°C for 30 seconds, 65°C for 30 seconds, and 68°C for 30 seconds; 68°C for 6 minutes. Amplified products were separated in 12% acrylamide gels (acrylamide/bisacrylamide, 29:1; Bio-Rad), stained with ethidium bromide, and visualized with a Ugenius camera (Syngene). Fragment identity was confirmed by blunt cloning into pBluescript II KS and sequencing.
To generate calibration curves, positive control genomic fragments containing an AATT site were amplified with a specific primer and one of the primers used for the test fragment: Myosin, F: CCCAAACCCTAACTTTTCTTTCTC, R: TGCTCGCCACAGCACAT; DHFR, F: AACTAAGGCATACTTTGCAGCAC, R: AATAACCTTATACTTACTCTGG; Cyclin D2, F: GACTGAGATTCTGCGGTTCC, R: TGGACGCGTCTCTCTCTTTC; and BCL6, F: CCGAAGATTAGTCCCACGTC, R: CCAAGCACTGTCTCGTCCTC.
For TP53, the test fragment was cloned into pBluescript II KS, and one of the GATT sites was mutated to AATT with a Quickchange XL site-directed mutagenesis kit (Stratagene) to serve as a template for the control genomic PCR. Positive control fragments were mixed with 500 ng of wild-type fragment at ratios of 5 × 10−3, 10−4, 10−5, and 10−6 and analyzed as described previously.
PhosTag SDS-PAGE and immunoblotting analysis
Various phosphorylated forms of Ube1 were separated in an SDS polyacrylamide gel copolymerized with a dinuclear Mn2+ complex of 1,3-bis[bis(pyridin-2-ylmethyl) amino]propan-2-olate (PhosTag-acrylamide; The PhosTag consortium). Cells were lysed in lysis buffer (20mM Tris-HCl, pH 8; 10% glycerol; and EDTA-free protease inhibitors cocktail; Sigma-Aldrich), phosphatase inhibitors cocktail (Sigma-Aldrich), and 2% Igepal (Sigma-Aldrich) protein concentration was determined by Bradford assay (Bio-Rad). Samples were run on 6% polyacrylamide midi-gels (acrylamide/bisacrylamide 29:1; Bio-Rad) containing 50μM PhosTag and 100μM MnCl2 at 5 mA per gel at room temperature. Gels were equilibrated in transfer buffer (192mM glycine; 25mM Tris-HCl, pH 8.6; 0.01% SDS; 15% methanol) supplemented with 1mM EDTA for 10 minutes, then without EDTA for 10 minutes. Proteins were transferred to polyvinylidene fluoride membranes (Bio-Rad) with a Criterion blotter (Bio-Rad) for 1 hour at 100 V. Membranes were blocked overnight with 0.5% BSA (Sigma-Aldrich), in TBST buffer (150mM NaCl; 50mM Tris-HCl, pH 7.4; 1% Tween-20; Acros), incubated with a polyclonal anti-Ube1 isoform E1A antibody (1:1000; Abcam) for 2 hours, followed by incubation with horseradish peroxidase-coupled antirabbit secondary antibody (Abcam, 1:10 000) for 1 hour. The blots were treated with ECL reagent (Amersham) and signal was detected with a LAS-3000 Imager (Fujifilm). To determine total Ube1 levels, cells were lysed in RIPA buffer (250mM NaCl, 2% Igepal, 0.5% sodium deoxycholate, 0.1% SDS, 50mM Tris-HCl, pH 8.0) and Western blotting was performed as described previously with 7.5%-12% gradient gels and a polyclonal anti-Ube1 antibody (isoforms E1A and E1B; Abcam) or a polyclonal anti-actin antibody (Sigma-Aldrich).
Results
Characterization of the system
The majority of circulating human B lymphocytes are quiescent cells of both naive and antigen-specific memory phenotype. Untouched quiescent B lymphocytes were isolated from PBMCs by negative selection with CD43-coated microbeads, then stained with antibodies to CD19 and IgD to assess purity (Figure 1A), and with DNA and RNA dyes to determine cell cycle profile (Figure 1A-B). The majority of purified B cells were CD19+IgD+, a characteristic of naive mature B cells, with a minor CD19+IgD− component typical of memory cells. As expected, all were in G0 phase, whereas the control cell lines, GM01953 (wild-type lymphoblasts) and GM01646 (NER-deficient lymphoblasts from a XP group E patient), displayed a standard cell-cycle profile.
Purification of B lymphocytes and proliferation after UV-irradiation. (A) Flow cytometric analysis of naive primary B lymphocytes stained with anti-CD19 PE and anti-IgD FITC antibodies to assess purity (left) and with propidium iodide for cell cycle profile (right). (B) Differential staining of DNA with Hoechst 33342 and RNA with Pyronin Y in naive primary B lymphocytes, wild-type (WT), and XP-E lymphoblasts. The percentage of cells in G0, G1, and G2 is indicated for each cell type. (C) Cell division analyses of CFSE-labeled primary B lymphocytes, WT, and XP-E lymphoblasts, after UV irradiation. Quiescent B lymphocytes were irradiated or not with 5 J/m2 254 nm UV light, pulse-labeled with the protein dye CFSE 24 hours later, and stimulated to proliferate with antihuman F(ab′)2 fragments and Pam3CSK4 lipopeptide (IP3). WT and XP-E lymphoblasts were similarly labeled with CFSE 24 hours after irradiation. Samples were analyzed 24 or 72 hours after labeling for the dilution of the CFSE signal resulting from cell division, excluding dead cells on the basis of propidium iodide staining. The shaded peak (CTRL) corresponds to the autofluorescence of unlabeled cells. Data are representative of 3 independent experiments. (D) Time course of [3H]thymidine incorporation in cells irradiated or not with 5 J/m2 UV light. After irradiation, quiescent B lymphocytes were cultured for 24 hours before stimulation. In all experiments, [3H]thymidine was added 20 hours before sample collection. Data are displayed as means of counts per minute (cpm) ± SEM of 2 independent experiments.
Purification of B lymphocytes and proliferation after UV-irradiation. (A) Flow cytometric analysis of naive primary B lymphocytes stained with anti-CD19 PE and anti-IgD FITC antibodies to assess purity (left) and with propidium iodide for cell cycle profile (right). (B) Differential staining of DNA with Hoechst 33342 and RNA with Pyronin Y in naive primary B lymphocytes, wild-type (WT), and XP-E lymphoblasts. The percentage of cells in G0, G1, and G2 is indicated for each cell type. (C) Cell division analyses of CFSE-labeled primary B lymphocytes, WT, and XP-E lymphoblasts, after UV irradiation. Quiescent B lymphocytes were irradiated or not with 5 J/m2 254 nm UV light, pulse-labeled with the protein dye CFSE 24 hours later, and stimulated to proliferate with antihuman F(ab′)2 fragments and Pam3CSK4 lipopeptide (IP3). WT and XP-E lymphoblasts were similarly labeled with CFSE 24 hours after irradiation. Samples were analyzed 24 or 72 hours after labeling for the dilution of the CFSE signal resulting from cell division, excluding dead cells on the basis of propidium iodide staining. The shaded peak (CTRL) corresponds to the autofluorescence of unlabeled cells. Data are representative of 3 independent experiments. (D) Time course of [3H]thymidine incorporation in cells irradiated or not with 5 J/m2 UV light. After irradiation, quiescent B lymphocytes were cultured for 24 hours before stimulation. In all experiments, [3H]thymidine was added 20 hours before sample collection. Data are displayed as means of counts per minute (cpm) ± SEM of 2 independent experiments.
Activation of human B cells requires two antigen-specific signals, one delivered by the antigen through the B-cell receptor (BCR), the second triggered by microbial products through Toll-like receptors.15 To induce proliferation, we thus stimulated B cells through BCR with IgG + IgM + IgA F(ab′) and with the bacterial lipopeptide Pam3CSK4, which is a Toll-like receptor 2 ligand. Proliferation was verified by CFSE dilution assay (Figure 1C) and by the measurement of [3H]thymidine incorporation during DNA replication (Figure 1D). B cells reached maximum proliferation 3 days after stimulation, at which time 4 peaks could be observed in their CFSE staining profile: undivided cells (33.3%), and cells having divided once (14.8%), twice (31.7%), or more (16.2%).
Because we intended to appraise NER by measuring the repair of UV-induced lesions, we also investigated the effect of irradiation with 254-nm UV-C light on B-cell proliferation. Although a dose of 10 J/m2 strongly inhibited proliferation (data not shown), B cells tolerated irradiation with 5 J/m2 UV (Figure 1C-D), and 36.5% of them had divided at least once 72 hours later.
Global genome repair is attenuated in quiescent cells
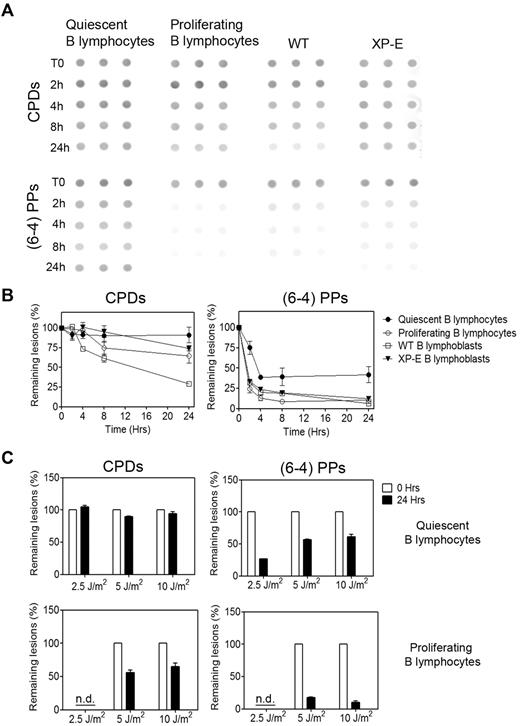
UV light induces two types of lesions, cyclobutane pyrimidine dimers (CPDs) and (6-4)pyrimidine-pyrimidone photoproducts, hereafter (6-4)PPs, which are repaired with very different kinetics6 ; repair of CPDs is slow and requires the specialized damaged DNA-binding enzyme DDB, whereas (6-4)PPs are repaired rapidly with no need for DDB. Quiescent B lymphocytes were irradiated with 5 J/m2 UV light, DNA isolated at various time points, and the amount of lesions measured by slot blot with mAbs specific for CPDs and (6-4)PPs (Cosmobio). Similar to what we previously reported for terminally differentiated cells,10,11 the repair of (6-4)PPs was unusually slow, and we could not detect any repair of CPDs within 24 hours (Figure 2). By contrast, B lymphocytes at the peak of their proliferation, 3 days after stimulation, proficiently repaired (6-4)PPs and displayed partial repair of CPDs, although not to the extent generally observed in other proliferating cells.
Quiescent B lymphocytes are deficient in global genome repair. (A) Time-course of repair of UV-induced lesions in quiescent and proliferating B lymphocytes and in WT and XP-E B lymphoblasts after exposure to 5 J/m2 UV light. DNA samples, 1 μg for (6-4)PPs and 60 ng for CPDs, were blotted in triplicate on nitrocellulose membranes, and the amount of lesions was visualized with anti-CPDs and anti-(6-4)PPs antibodies. (B) Quantification of at least 3 experiments, including the aforementioned one, with ImageGauge software. To control for variations in DNA load, blots were reprobed with a 32P-labeled genomic probe, and the antibody signal was normalized against genomic DNA. Data are shown as mean ± SEM. The difference in repair efficiencies between quiescent and proliferating B lymphocytes was statistically significant as determined by paired Student t test, P < .05, n = 30 donors. (C) Repair of CPDs and (6-4)PPs lesions in quiescent (top) and proliferating B lymphocytes (bottom) at 24 hours after exposure to various doses of UV. Data are representative of 2 experiments and are shown as mean ± SEM. The difference in repair efficiencies between quiescent and proliferating B lymphocytes was statistically significant as determined by paired Student t test, P < .05, n = 6 donors.
Quiescent B lymphocytes are deficient in global genome repair. (A) Time-course of repair of UV-induced lesions in quiescent and proliferating B lymphocytes and in WT and XP-E B lymphoblasts after exposure to 5 J/m2 UV light. DNA samples, 1 μg for (6-4)PPs and 60 ng for CPDs, were blotted in triplicate on nitrocellulose membranes, and the amount of lesions was visualized with anti-CPDs and anti-(6-4)PPs antibodies. (B) Quantification of at least 3 experiments, including the aforementioned one, with ImageGauge software. To control for variations in DNA load, blots were reprobed with a 32P-labeled genomic probe, and the antibody signal was normalized against genomic DNA. Data are shown as mean ± SEM. The difference in repair efficiencies between quiescent and proliferating B lymphocytes was statistically significant as determined by paired Student t test, P < .05, n = 30 donors. (C) Repair of CPDs and (6-4)PPs lesions in quiescent (top) and proliferating B lymphocytes (bottom) at 24 hours after exposure to various doses of UV. Data are representative of 2 experiments and are shown as mean ± SEM. The difference in repair efficiencies between quiescent and proliferating B lymphocytes was statistically significant as determined by paired Student t test, P < .05, n = 6 donors.
As expected, wild-type lymphoblasts proficiently repaired both CPDs and (6-4)PPs, the latter faster than the former. XP-E lymphoblasts, lacking the XPE/DDB2 subunit of the damage-recognition enzyme DDB,16 were deficient in CPD repair at the global genome level but retained proficient repair of (6-4)PPs, for which DDB is not required.
Active genes remain proficiently repaired by DAR and TCR
Both types of UV-induced lesions are known to stall RNA polymerase II,17 and the accumulation of UV-induced damage in transcribed genes would thus have severe consequences for cell survival if left unrepaired. We have previously observed that NER remains proficient in active genes in terminally differentiated cells that experience a marked down-regulation in NER at the global genome level,10 and we set out to verify whether it was also the case for quiescent B lymphocytes.
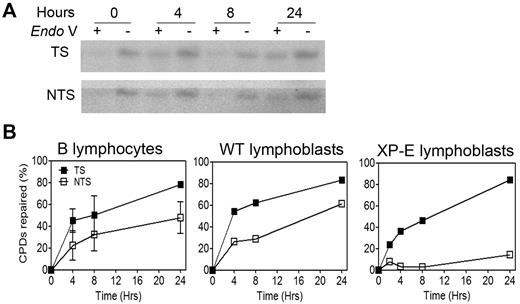
Repair of CPD lesions in the constitutively expressed dihydrofolate reductase (DHFR) gene was appraised by a modified Southern blot technique in which DNA is treated with T4 endonuclease V, an enzyme that nicks the DNA backbone next to CPDs. The use of strand-specific RNA probes then allows to monitor repair of CPDs over time in a gene-specific and strand-specific manner.13 Because TCR relies on RNA polymerase II for lesion detection, it is not impaired by the lack of DDB in XP-E cells, which retain proficient repair of CPDs in the transcribed-strand of active genes.16 As a result, XP-E cells tolerate the level of UV irradiation required by our experiments, which would likely be lethal to other XP cells. In accordance with these previously published data,16 we observed virtually no repair of the nontranscribed strand in XP-E cells, whereas repair of the transcribed strand was normal (Figure 3B). By contrast, both strands were proficiently repaired in quiescent B lymphocytes (Figure 3A-B), despite their complete lack of CPD repair at the global genome level (Figure 2). Proficient repair of the nontranscribed strand in these cells constitutes DAR, whereas the repair bias in favor of the transcribed strand is the hallmark of TCR.
Repair in a constitutively expressed gene remains proficient in quiescent B lymphocytes. (A) Representative experiment (n = 10 donors) showing repair of CPD lesions in B lymphocytes in the transcribed (TS) and nontranscribed (NTS) strands of the dihydrofolate reductase gene. Cells were harvested at various times after irradiation with 10 J/m2 UV light. For each time point, 50 μg of genomic DNA was digested with KpnI, half of the sample was treated (+) or not treated (−) with the CPD-specific nicking enzyme T4 endonuclease V (Endo V), resolved in a denaturing agarose gel, transferred to a nylon membrane, and hybridized with 32P-labeled strand-specific RNA probes. (B) Quantification of 2 independent experiments, including the aforementioned one. WT and XP-E B lymphoblasts are included for reference. The amount of lesion-free fragment was quantified with Molecular Analyst software, and lesion frequency was determined with the Poisson expression. Error bars represent SEM.
Repair in a constitutively expressed gene remains proficient in quiescent B lymphocytes. (A) Representative experiment (n = 10 donors) showing repair of CPD lesions in B lymphocytes in the transcribed (TS) and nontranscribed (NTS) strands of the dihydrofolate reductase gene. Cells were harvested at various times after irradiation with 10 J/m2 UV light. For each time point, 50 μg of genomic DNA was digested with KpnI, half of the sample was treated (+) or not treated (−) with the CPD-specific nicking enzyme T4 endonuclease V (Endo V), resolved in a denaturing agarose gel, transferred to a nylon membrane, and hybridized with 32P-labeled strand-specific RNA probes. (B) Quantification of 2 independent experiments, including the aforementioned one. WT and XP-E B lymphoblasts are included for reference. The amount of lesion-free fragment was quantified with Molecular Analyst software, and lesion frequency was determined with the Poisson expression. Error bars represent SEM.
Mechanism of NER attenuation in quiescent B cells
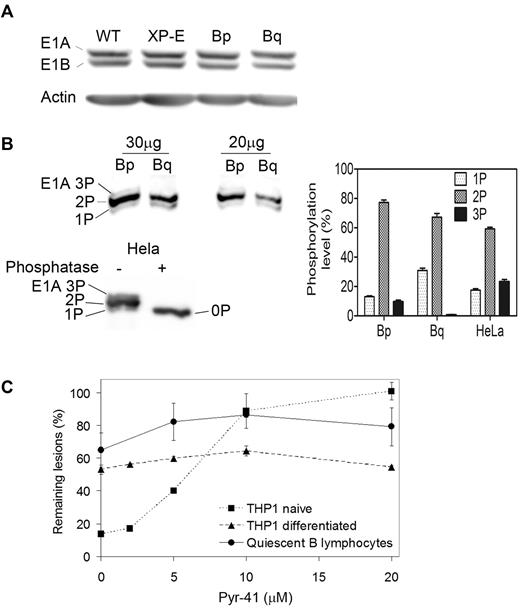
Because the repair phenotype of quiescent B lymphocytes closely matches that of terminally differentiated cells, we suspected that it may stem from the same mechanism, driven by modulations in Ube1 phosphorylation. Indeed, we found no difference in the amount of Ube1 in quiescent or proliferating B cells (Figure 4A) but observed approximately 20% less phosphorylation in quiescent cells (Figure 4B), with a decrease in multiphosphorylated forms (3P, 2P) and a consequential increase in mono-phosphorylated Ube1 (1P). This finding is similar to what we observed on macrophage differentiation by using metabolic labeling,9 2D-gel electrophoresis,9 or Phos-tag gel retardation (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Interestingly, 3 days after stimulation, phosphorylation of Ube1 in proliferating B lymphocytes had reverted to the pattern observed with purified Ube1, isolated from repair-competent HeLa cells (Figure 4B, bottom). Thus the reduction in Ube1 phosphorylation in quiescent B cells correlated with the NER impairment and so did the concurrent increase in Ube1 phosphorylation and recovery of NER upon proliferation.
Ube1 phosphorylation and impact on NER in quiescent B lymphocytes. (A) Representative immunoblot showing the amount of ubiquitin-activating enzyme Ube1 in 50 μg of total cell lysate from wild-type (WT) or XP-E lymphoblasts, and from proliferating (Bp) or quiescent (Bq) B lymphocytes (n = 20 donors). Western blotting was performed with an antibody directed against both the E1A (full length, phosphorylated) and E1B (truncated) isoforms of Ube1. Beta-actin was used as a loading control. (B) Top, Mobility shift detection of phosphorylated forms of Ube1 isoform E1A, 3 phosphate (3P), 2 phosphate (2P), and 1 phosphate (1P) in proliferating and quiescent B lymphocytes. Cell lysates were run on a 6% SDS-PAGE containing 50μM of acrylamide-coupled phosphate chelator (PhosTag) and analyzed by Western blotting with an antibody specific for the E1A isoform. Bottom, His-tagged Ube1 was purified from Hela cells, treated or not treated with λ phosphatase, and analyzed as described previously. The phosphatase-treated sample reveals the position of nonphosphorylated Ube1 (0P). Right, Quantification of the phosphorylated forms of Ube1 isoform E1A in proliferating and quiescent B lymphocytes and of purified Ube1 with ImageGauge software. Data are representative of 3 experiments and shown as mean ± SEM. (C) Effect of the Ube1-specific inhibitor Pyr-41 on repair of (6-4)PPs at 3 hours after irradiation in naive and differentiated THP1 leukemia cells, and in quiescent B lymphocytes (n = 6 donors).
Ube1 phosphorylation and impact on NER in quiescent B lymphocytes. (A) Representative immunoblot showing the amount of ubiquitin-activating enzyme Ube1 in 50 μg of total cell lysate from wild-type (WT) or XP-E lymphoblasts, and from proliferating (Bp) or quiescent (Bq) B lymphocytes (n = 20 donors). Western blotting was performed with an antibody directed against both the E1A (full length, phosphorylated) and E1B (truncated) isoforms of Ube1. Beta-actin was used as a loading control. (B) Top, Mobility shift detection of phosphorylated forms of Ube1 isoform E1A, 3 phosphate (3P), 2 phosphate (2P), and 1 phosphate (1P) in proliferating and quiescent B lymphocytes. Cell lysates were run on a 6% SDS-PAGE containing 50μM of acrylamide-coupled phosphate chelator (PhosTag) and analyzed by Western blotting with an antibody specific for the E1A isoform. Bottom, His-tagged Ube1 was purified from Hela cells, treated or not treated with λ phosphatase, and analyzed as described previously. The phosphatase-treated sample reveals the position of nonphosphorylated Ube1 (0P). Right, Quantification of the phosphorylated forms of Ube1 isoform E1A in proliferating and quiescent B lymphocytes and of purified Ube1 with ImageGauge software. Data are representative of 3 experiments and shown as mean ± SEM. (C) Effect of the Ube1-specific inhibitor Pyr-41 on repair of (6-4)PPs at 3 hours after irradiation in naive and differentiated THP1 leukemia cells, and in quiescent B lymphocytes (n = 6 donors).
To determine the importance of Ube1 for NER efficiency during quiescence, we challenged quiescent B cells with Pyr-41, an Ube1-specific inhibitor.18 Pyr-41 inhibits NER in a dose-dependent manner in proliferating cells but does not further impair the already partially deficient repair of (6-4)PP lesions in differentiated cells, likely because the incomplete phosphorylation of Ube1 in the latter prevents it from activating NER anyway.9 The effect of Pyr-41 in quiescent B lymphocytes mirrored that in differentiated cells (Figure 4C). Taken together, these observations strongly suggest that the down-regulation of NER in quiescent B cells proceeds from the same mechanism as in differentiated cells.
Proliferation results in high mutagenesis in previously silent genes
Contrarily to differentiated cells, though, UV-irradiated B lymphocytes retain the ability to proliferate, and the accumulation of unrepaired DNA lesions is likely to result in high mutagenesis during DNA replication. Measuring it with the fluctuation assay19 was not feasible because of the limited proliferative potential of B lymphocytes and because the active HPRT gene ought to be proficiently repaired. We thus adapted a PCR technique that is applicable to any genomic location and relies on the creation of a new restriction site by a mutation.14 During replication, CPDs are generally bypassed by translesion polymerase eta, which tends to insert adenine opposite the 3′ cytosine in the dimer, giving rise to a TC to TT mutation. The contribution of (6-4)PPs to mutagenesis is probably minor, as these are induced at much lower levels than CPDs (approximately in a 1:4 ratio) and substantially repaired, even in quiescent cells. Genomic fragments containing AATC sequences were preamplified with high-fidelity polymerase and digested with the restriction enzyme Tsp509I, which recognizes AATT and thus only cuts mutated products. A linker with an AATT cohesive end was then ligated to the restricted products, and two rounds of nested PCR performed with linker-specific primers, so as to selectively amplify mutated fragments (Figure 5A). This technique differs from the “restriction site mutation” assay pioneered by Parry et al,20 because it detects mutations that create a restriction site rather than destroy it, and thus does not experience the known drawbacks of restriction site mutation. In addition, because the wild-type sequence is not palindromic, we can select potential mutation sites on either the transcribed (GATT) or nontranscribed (AATC) strand. To determine mutation frequency, we constructed standard curves by spiking the genomic PCR with minute amounts of positive control, generated either by site-directed mutagenesis or by amplifying a slightly longer genomic fragment encompassing an AATT site. We verified that our technique allows to measure ratios as low as 10−6.
UV-induced mutagenesis in silent and constitutively expressed genes. (A) Schematic representation of the strategy used to appraise UV-induced mutagenesis. Genomic fragments from the indicated genes were chosen to contain the possible mutation sites GATT (T <> C) dimer in the transcribed strand) or AATC (dimer in the nontranscribed stand), both converted to AATT if a mutation occurs during replication, allowing digestion with Tsp509I. Nucleotide numbering is according to NCBI. Fragment sizes (after digestion, ligation of an adapter, and amplification by nested PCR) are shown next to each potential mutation site. For positive controls, longer sequences (black bars) containing a genuine AATT site were amplified with a different primer. For TP53, the positive control was created by site-directed mutagenesis of the downstream GATT site. (B) Mutagenesis in the silent Myosin gene, in cells irradiated or not irradiated with 5 J/m2 UV. Top left, A 98-bp mutated fragment was detected in all 3 cell types, and induced by UV in B lymphocytes (n = 12 donors) and XP-E lymphoblasts. Bottom left, Calibration curve used to appraise mutation frequency. Amplification of a 112-bp control band was detected after diluting the positive control genomic fragment with the test fragment down to a ratio of 5 × 10−6. Right, After quantification of ethidium bromide fluorescence, the mutation frequency in each cell type was determined on the basis of the calibration curve. Data are representative of 3 distinct experiments and shown as mean ± SEM. (C) Mutagenesis in the nontranscribed strand (NTS) of the DHFR gene. Top left, 2 mutated fragments of 186 bp and 113 bp were detected in UV-irradiated XP-E B lymphoblasts but not in WT lymphoblasts or B lymphocytes (n = 10 donors). Bottom left, Calibration curve for the DHFR control fragment. Right, Quantification of mutation frequency based on the calibration curve. Data are representative of 3 distinct experiments, and shown as mean ± SEM. (D) Mutagenesis in the transcribed strand (TS) of the TP53 gene. Top left, Amplification of a 265-bp and an 89-bp fragment was expected if a mutation occurred, but only background levels were detected, irrespective of UV irradiation. Bottom left, Calibration curve for the TP53 control fragment. This control, generated by site-directed mutagenesis, has the same size (265 bp) as the expected fragment and is detectable down to a dilution of 5 × 10−6. Right, Quantification of mutation frequency in the TP53 fragment.
UV-induced mutagenesis in silent and constitutively expressed genes. (A) Schematic representation of the strategy used to appraise UV-induced mutagenesis. Genomic fragments from the indicated genes were chosen to contain the possible mutation sites GATT (T <> C) dimer in the transcribed strand) or AATC (dimer in the nontranscribed stand), both converted to AATT if a mutation occurs during replication, allowing digestion with Tsp509I. Nucleotide numbering is according to NCBI. Fragment sizes (after digestion, ligation of an adapter, and amplification by nested PCR) are shown next to each potential mutation site. For positive controls, longer sequences (black bars) containing a genuine AATT site were amplified with a different primer. For TP53, the positive control was created by site-directed mutagenesis of the downstream GATT site. (B) Mutagenesis in the silent Myosin gene, in cells irradiated or not irradiated with 5 J/m2 UV. Top left, A 98-bp mutated fragment was detected in all 3 cell types, and induced by UV in B lymphocytes (n = 12 donors) and XP-E lymphoblasts. Bottom left, Calibration curve used to appraise mutation frequency. Amplification of a 112-bp control band was detected after diluting the positive control genomic fragment with the test fragment down to a ratio of 5 × 10−6. Right, After quantification of ethidium bromide fluorescence, the mutation frequency in each cell type was determined on the basis of the calibration curve. Data are representative of 3 distinct experiments and shown as mean ± SEM. (C) Mutagenesis in the nontranscribed strand (NTS) of the DHFR gene. Top left, 2 mutated fragments of 186 bp and 113 bp were detected in UV-irradiated XP-E B lymphoblasts but not in WT lymphoblasts or B lymphocytes (n = 10 donors). Bottom left, Calibration curve for the DHFR control fragment. Right, Quantification of mutation frequency based on the calibration curve. Data are representative of 3 distinct experiments, and shown as mean ± SEM. (D) Mutagenesis in the transcribed strand (TS) of the TP53 gene. Top left, Amplification of a 265-bp and an 89-bp fragment was expected if a mutation occurred, but only background levels were detected, irrespective of UV irradiation. Bottom left, Calibration curve for the TP53 control fragment. This control, generated by site-directed mutagenesis, has the same size (265 bp) as the expected fragment and is detectable down to a dilution of 5 × 10−6. Right, Quantification of mutation frequency in the TP53 fragment.
Quiescent B lymphocytes were irradiated with 5 J/m2 UV, stimulated 24 hours later, and harvested after 3 days of proliferation. We first measured mutagenesis in the Myosin gene, which is silent in B lymphocytes and should thus accumulate CPDs. Indeed, we found that mutation frequency in B lymphocytes was greater than in wild-type cells, similar to that of NER-deficient XP-E lymphoblasts (Figure 5B). By contrast, the mutation frequency in two constitutively expressed genes, DHFR (Figure 5C) and TP53 (Figure 5D), was below detection level in B lymphocytes. In XP-E cells, the mutation frequency was elevated in the nontranscribed strand and undetectable in the transcribed strand, illustrating the fact that TCR is proficient in XP-E cells.
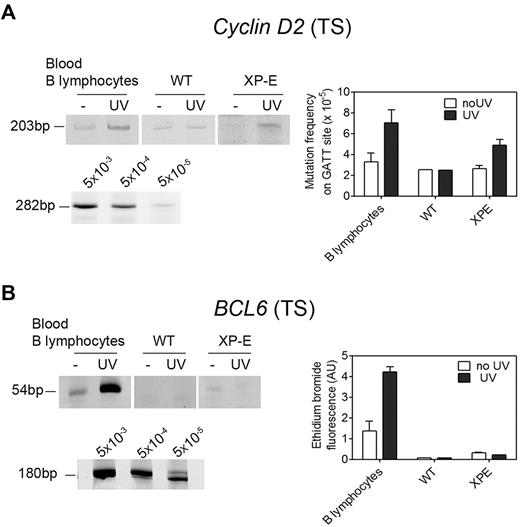
More interesting to us were genes that are silent in quiescent cells, thereby accumulating damage, but become active in proliferating cells. After activation they will be repaired by DAR and TCR, but these processes may not be fast enough to prevent mutagenesis. We first examined the Cyclin D2 gene, which is rapidly induced via BCR stimulation as cells transit from G0 to G1.21 Most interestingly, B lymphocytes displayed an elevated mutation frequency in this gene (Figure 6A), indicating that recovery of NER on proliferation was too slow to efficiently clean up a previously silent gene before it is replicated. The same was true for the BCL6 gene, which is silent in naive B lymphocytes and induced on proliferation in germinal center B cells. We observed strong UV-induced mutagenesis in the 5′ noncoding region of BCL6 in B lymphocytes (Figure 6B), at a site that was found mutated in lymphoma patients.22
UV-induced mutagenesis in inducible genes. (A) Mutagenesis in the transcribed strand of the Cyclin D2 gene. Top left, Detection of a mutated fragment of 203 bp in nonirradiated and UV-irradiated cells. Bottom left, Calibration curve for the 282-bp control fragment. Right, Quantification of mutation frequency on the basis of the calibration curve. Data are representative of 3 distinct experiments (n = 12 donors) and shown as mean ± SEM. (B) Mutagenesis in the transcribed strand of the BCL6 gene in cells irradiated or not irradiated with 5 J/m2 UV. Top left, A 54-bp mutated fragment was strongly amplified in UV-irradiated primary B lymphocytes (n = 10 donors) but not in WT or XP-E lymphoblasts. Bottom left, Calibration curve for the 180-bp control fragment. Right, Densitometric quantification of ethidium bromide-stained bands. Mutation frequency was not calculated because of the significant difference in sizes between the mutated fragment and the positive control. Data are representative of 3 distinct experiments, and shown as mean ± SEM.
UV-induced mutagenesis in inducible genes. (A) Mutagenesis in the transcribed strand of the Cyclin D2 gene. Top left, Detection of a mutated fragment of 203 bp in nonirradiated and UV-irradiated cells. Bottom left, Calibration curve for the 282-bp control fragment. Right, Quantification of mutation frequency on the basis of the calibration curve. Data are representative of 3 distinct experiments (n = 12 donors) and shown as mean ± SEM. (B) Mutagenesis in the transcribed strand of the BCL6 gene in cells irradiated or not irradiated with 5 J/m2 UV. Top left, A 54-bp mutated fragment was strongly amplified in UV-irradiated primary B lymphocytes (n = 10 donors) but not in WT or XP-E lymphoblasts. Bottom left, Calibration curve for the 180-bp control fragment. Right, Densitometric quantification of ethidium bromide-stained bands. Mutation frequency was not calculated because of the significant difference in sizes between the mutated fragment and the positive control. Data are representative of 3 distinct experiments, and shown as mean ± SEM.
Discussion
In this study, we demonstrate for the first time the direct role of UV radiation in induction of point mutations in primary B lymphocytes, a process that can contributes to the genesis of lymphoma in vivo. Indeed, strong exposure to UV radiation is an important environmental risk factor for non-Hodgkin lymphoma. Epidemiologic studies have established a shared etiology between non-Hodgkin lymphoma and skin cancer, both being positively associated with the levels of solar UV radiation.23,24
But, can UV fluxes directly reach circulating B lymphocytes in vivo? Evidence supports the concept that strong exposure to UV radiation triggers DNA damage in circulating immune cells, drastically affecting their biologic functions in vivo. UV-induced DNA lesions, and particularly pyrimidine dimers, were shown to initiate the development of erythema and immune suppression.25,26 It is now well accepted that exposure to UV radiation results in systemic immune suppression, which is of hematopoetic origin. Very interestingly, splenocytes from UV-irradiated mice transfer systemic immune suppression in syngeneic nonirradiated mice, in a model of delayed type hypersensitivity response, and this process is inhibited by T4 endonuclease V-containing liposomes that specifically repair pyrimidine dimers.27 Moreover, UV radiation affects both cellular and humoral immune responses and sometimes triggers a switch from cellular to humoral immunity, proving that it affects B lymphocyte functions in vivo.28
Our results, together with previous studies,29-31 establish that peripheral blood lymphocytes are deficient in global genome repair when quiescent but can regain some NER activity on proliferation. Those studies, however, were not performed on a pure B lymphocyte population and only considered NER at the global genome level. By contrast, we found that NER remains fully proficient in an active gene, in which both strands were proficiently repaired; the transcribed strand better than the nontranscribed strand. This strand bias indicates that TCR is proficient,6 whereas the proficient repair of the nontranscribed strand in otherwise repair-deficient cells constitutes DAR.10
This peculiar repair phenotype mirrors exactly that observed in differentiated cells.8 We have previously demonstrated that NER down-regulation in macrophages ultimately results from reduced phosphorylation of the ubiquitin-activating enzyme Ube1.9 Here we observed a similar reduction in Ube1 phosphorylation in quiescent B cells, correlating with the NER impairment. As in differentiated cells, inhibition of Ube1 with Pyr-41 had little effect on NER in quiescent B cells, suggesting that the impairment of NER in B cells proceeds from the same mechanism as in differentiated cells. The determination of which residue on Ube1 is dephosphorylated during quiescence/differentiation and its importance for proficient NER (and possibly other pathways controlled by ubiquitination) are important issues that remain to be resolved. We have undertaken a systematic identification of phosphorylation sites on Ube1 (21 so far) and of their influence on the charging of ubiquitin on the various E2 enzymes. We have preliminary evidence that minute changes in Ube1 phosphorylation can produce a significant alteration in the rate of ubiquitin charging on a given E2 without affecting others (data not shown). This may account for the selectivity of the proposed mechanism, which can limit ubiquitination of an NER enzyme whereas ubiquitination as a whole remains proficient.
UV-irradiated B lymphocytes retain the ability to proliferate, and accumulated DNA lesions cause a high rate of mutagenesis during DNA replication. By using a restriction digest-PCR method, we found that UV-induced mutagenesis in a silent gene was as high in B lymphocytes as in NER-deficient cells. By calibration against a standard curve, we estimated a mutation frequency in the order of 10−4, which correlates well with the amount of UV-induced damage and the error rate of polymerase eta,32,33 as well as with frequencies observed with the fluctuation assay.34 As expected because NER remains proficient in constitutively expressed genes, we did not detect UV-induced increases in mutation frequency in the active DHFR and TP53 genes in quiescent B lymphocytes. Conceivably, mutations confined to silent genes might have little consequences for the host, although it is possible that a promoter mutation could activate a proto-oncogene. Indeed, expression of genes specific for common lymphoid precursors, and inappropriate for B-cell lineage, has been reported in Hodgkin lymphoma.35 A classic example is the abnormal expression of ζ-associated protein of 70 kDa (ZAP70) in B cells of chronic lymphocytic leukemia patients.36
Even more interesting is the case of genes that are silent in quiescent cells, thereby accumulating damage, but become active in proliferating cells. After activation they will be repaired by DAR and TCR, but we observed that these processes are not fast enough to prevent mutagenesis. We examined the Cyclin D2 gene, which is rapidly induced via BCR stimulation as cells transit from G0 to G1,21 and found that mutagenesis is almost as high as in a silent gene, with frequencies similar to those of repair-deficient cells. This phenomenon may have ominous consequences, as mutations in cell-cycle inducible genes, leading to deregulation of their expression and function, are usually associated with malignant transformation. Interestingly, cyclin D2 is constitutively expressed at high levels in B-cell lymphoma.37
Point mutations in several genes, including c-Myc,38 TP53,39 BRAF,40 BCL-2,41 BCL-X,42 and BCL622,43 have been implicated in the pathogenesis of B lymphoma. BCL6, a transcriptional repressor of B-cell terminal differentiation, is silent in naive B lymphocytes and induced upon proliferation in germinal center B cells. Furthermore, memory B cells maintain expression of BCL6 to retain their self-renewal potential.44,45 Point mutations in BCL6 are documented in many lymphomas22 and are mainly located within a hotspot in the 5′ untranslated region.46 Strikingly, we observed strong UV-induced mutagenesis in this region of BCL6 in B lymphocytes. Interestingly, Cyclin D2 expression can be regulated by BCL6.47 Therefore, a combination of mutagenic events targeting both Cyclin D2 and BCL6, as shown here, singles out the NER-deficient quiescent B cells as a possible cellular origin for B-cell malignancies.
NER attenuation during quiescence and somatic hypermutation occurring during B-cell activation both introduce mutations in B cells genome via error-prone translesion synthesis. By itself, a deficiency in NER is unlikely to directly affect somatic hypermutation in B lymphocytes, because cells from patients with inherited defects of NER show no defect in this process nor in rearrangement of Ig genes.48,49 However, hereditary defects in NER can result in the initiation of B-cell malignancies or abnormal immune functions.7
In conclusion, down-regulation of NER appears to be a common feature of G0-arrested cells, whether quiescent or terminally differentiated, and predisposes them to genomic instability. The majority of circulating B lymphocytes are quiescent, and their NER impairment means that they likely accumulate DNA damage from various sources besides UV, such a chemicals from tobacco smoke, atmospheric pollution, or food carcinogens. When these cells receive the appropriate proliferative stimuli, eg, during an infection, they will accumulate potentially deleterious mutations. Strikingly, the overwhelming majority of B lymphomas originate from cells that underwent antigenic stimulation,50 in full accordance with our model. Thus, we propose that NER down-regulation in quiescent B lymphocytes predisposes them to increased mutagenesis, potentially perturbing gene expression and favoring aberrant rearrangements, with important implications for cell replication and neoplastic transformation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Stephen Lloyd for the gift of T4 endonuclease V and Mark Meuth and Tomas Lindahl for their critical reading of the manuscript.
This work was supported by grants from Leukemia and Lymphoma Research United Kingdom (06049) and the United Kingdom Biotechnology and Biological Sciences Research Council (BB/E006590/1).
Authorship
Contribution: N.H.-N. and T.N. designed the experiments; N.H.-N. performed all experiments except for those in Figure 4, which were performed by K.L. and W.-T.L.; and N.H.-N. and T.N. wrote the paper.
The current affiliation for W.-T.L. is MRC Toxicology unit, University of Leicester, Leicester, United Kingdom. The current affiliation for K.L. is Strathclyde Institute of Pharmacy and Biomedical Sciences, University of Strathclyde, Glasgow, United Kingdom.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Thierry Nouspikel, Institute for Cancer Studies, University of Sheffield, Sheffield, S10 2RX, United Kingdom; e-mail: t.nouspikel@sheffield.ac.uk.

![Figure 1. Purification of B lymphocytes and proliferation after UV-irradiation. (A) Flow cytometric analysis of naive primary B lymphocytes stained with anti-CD19 PE and anti-IgD FITC antibodies to assess purity (left) and with propidium iodide for cell cycle profile (right). (B) Differential staining of DNA with Hoechst 33342 and RNA with Pyronin Y in naive primary B lymphocytes, wild-type (WT), and XP-E lymphoblasts. The percentage of cells in G0, G1, and G2 is indicated for each cell type. (C) Cell division analyses of CFSE-labeled primary B lymphocytes, WT, and XP-E lymphoblasts, after UV irradiation. Quiescent B lymphocytes were irradiated or not with 5 J/m2 254 nm UV light, pulse-labeled with the protein dye CFSE 24 hours later, and stimulated to proliferate with antihuman F(ab′)2 fragments and Pam3CSK4 lipopeptide (IP3). WT and XP-E lymphoblasts were similarly labeled with CFSE 24 hours after irradiation. Samples were analyzed 24 or 72 hours after labeling for the dilution of the CFSE signal resulting from cell division, excluding dead cells on the basis of propidium iodide staining. The shaded peak (CTRL) corresponds to the autofluorescence of unlabeled cells. Data are representative of 3 independent experiments. (D) Time course of [3H]thymidine incorporation in cells irradiated or not with 5 J/m2 UV light. After irradiation, quiescent B lymphocytes were cultured for 24 hours before stimulation. In all experiments, [3H]thymidine was added 20 hours before sample collection. Data are displayed as means of counts per minute (cpm) ± SEM of 2 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/23/10.1182_blood-2010-12-326637/4/m_zh89991172270001.jpeg?Expires=1763513857&Signature=zG8Eze1USk3uk3Od1Ngu8lWEs4xElET8iA14zG-kef6UIagJQ5C750HwPA3nQrVQuzSeHvd8Wrg78Vsdn26lb4PFAI5wwDuxAqr-2wVDC-5U~Lw3fpiyZhH32vpM2v6O2KnE9t15QZ6G7oWUg1J~bBKKx9dPqZTfLXupWP0VE9ChMKviyvegE3oLxKlppL8g1Xg685a7UAjra2Q~eGgB9KUkdgjfjCUDK3uYxie6NXA2oq82TbzGulQlkuoG0b6pciobHPA4pIaGGV-29UbHDyy8LOghU5plhsJo9BA2SLMMdDigXPTEAnTUiBCgHqFruCGUmWMq7cPVXywC0cpnDw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal