Abstract
The molecular mechanisms regulating self-renewal of leukemia stem cells remain poorly understood. Here we report the generation of 2 closely related leukemias created through the retroviral overexpression of Meis1 and Hoxa9. Despite their apparent common origin, these clonal leukemias exhibit enormous differences in stem cell frequency (from 1 in 1.4, FLA2; to 1 in 347, FLB1), suggesting that one of these leukemias undergoes nearly unlimited self-renewal divisions. Using next-generation RNA-sequencing, we characterized the transcriptomes of these phenotypically similar, but biologically distinct, leukemias, identifying hundreds of differentially expressed genes and a large number of structural differences (eg, alternative splicing and promoter usage). Focusing on ligand-receptor pairs, we observed high expression levels of Sdf1-Cxcr4; Jagged2-Notch2/1; Osm-Gp130; Scf-cKit; and Bmp15-Tgfb1/2. Interestingly, the integrin beta 2-like gene (Itgb2l) is both highly expressed and differentially expressed between our 2 leukemias (∼ 14-fold higher in FLA2 than FLB1). In addition, gene ontology analysis indicated G-protein-coupled receptor had a much higher proportion of differential expression (22%) compared with other classes (∼ 5%), suggesting a potential role regulating subtle changes in cellular behavior. These results provide the first comprehensive transcriptome analysis of a leukemia stem cell and document an unexpected level of transcriptome variation between phenotypically similar leukemic cells.
Introduction
The ability of a distinct population of cells, termed stem cells, to maintain pluripotency is the foundation for the development of complex multicellular life. Within mammals, such multipotent cells are required to maintain a homeostatic blood supply, with a full complement of various terminally differentiated cell types.1 Despite the importance of this process, the molecular mechanisms underlying the maintenance of stem cells is still poorly understood. It is clear, however, that in many cases the neoplastic transformation of blood cells often involves either the failure or reversal of termination differentiation, resulting in primitive “cancer stem cells” (CSCs), which divide in an unregulated manner.2-4 CSCs are best described in humans, in which the rare so-called leukemia stem cells (leukemic hematopoietic stem cells [L-HSCs]) can be prospectively isolated and shown to transmit the disease when introduced into immunocompromised mice.5 Cells that do not share this phenotype often represent the bulk of the leukemic clone but fail to transmit the disease on transplantation.
In syngeneic mouse models of leukemia, L-HSC frequencies have been found to vary considerably and, in some cases, to be further enriched in fractions of the bulk population that express markers typically associated with normal HSCs. Neering et al,6 for example, reported that BCR-ABL + NUP98-HOXA9-induced mouse acute myeloid leukemia (AML) show a very low frequency of L-HSCs, which can be enriched to 1 in 7 cells by sorting for the lineage-negative Sca1+ FLT3+ population. In other murine AML models initiated by different oncogenic lesions, L-HSCs can be observed over a much wider range of frequencies, for example, 0.003% for Pten−/− AML,7 1% in E2A-PBX1 + Hoxa9 AML,8 and 2.3% to 71.4% for several Hoxa9 + Meis1-induced leukemias (current article).
Genes that are functionally significant for L-HSC expansion may represent the ultimate therapeutic targets for the disease. Such genes may be specific to the molecular alterations that initiate or sustain these diseases. For example, Meis1, a cofactor to Hoxa9 in AML, was recently reported as an important determinant of L-HSC frequency in MLL-AF10- and MLL-AF4-induced leukemias.9 Similarly, shRNA targeting of the myocyte enhancer factor 2 gene c, expressed at higher levels in L-HSCs of MLL-AF9 leukemias, decreased the frequency of L-HSCs by more than 50%.10 The polycomb group gene Bmi1 is also required for L-HSC self-renewal in Hoxa9 + Meis1-induced leukemias.10
In this paper, we describe the generation of a series of Hoxa9 + Meis1 AML derived from purified fetal liver (FL) cells. As detailed below, all these normal karyotype leukemias are remarkably similar in several aspects, including their frequency of L-HSCs (typically between 1 in 30-44) except for one leukemia (FLA2) in which almost each cell is an L-HSC. Given the possibility that differences in CSC determinates may be reflected in the transcriptomes of these 2 leukemias, we performed RNA-seq11,12 to characterize the respective cell types using a SOLiD next generation sequencer, in addition to analyzing the same RNA samples on Nimblegen expression microarrays. We generated 9 or 10 Gbp (Gigabase pairs) worth of mapped, strand-specific, sequence per cell type from 2 biologic replicates (individually grafted mice). Detailed analysis of these sequencing data revealed that a large number of genes are differentially expressed, including virtually all genes seen to be differentially regulated by microarray analysis. The RNA-seq data also revealed a surprisingly large number of structural variations in the transcriptomes of the 2 leukemias that hold the potential to substantially alter the proteome content between the leukemias. This study clearly demonstrates that even very closely related leukemias may differ substantially at the level of transcriptome diversity.
Methods
Mice
Donor C57BL/6:Pep3b (Ly5.1) and recipient C57BL/6 (Ly5.2) mice were bred and maintained as described.13 Experimental procedures were revised and approved by the University of Montreal animal ethics committee (Comité de Déontologie de l'Expérimentation sur les Animaux de l'Université de Montréal).
Isolation of murine hematopoietic cell subpopulations
Cells were first purified for Sca1 using magnetic activated cell sorting as per the manufacturer's instructions (Miltenyi Biotec). Further antibody staining and sorting to purify the Kit+, Lineage− (CD45R−, CD3−, Gr-1−, Ter119−) population were performed as described.14
Retroviral infection and transplantation
Infection and transplantation procedures were performed as described.15 Briefly, Hoxa9-ires-Meis1-pgk-Neo retroviral vectors were constructed, and high-titer viral producers were generated in the GPE-86 packaging line. FL cells obtained from (PepC3)F1(Ly5.1) mice were cocultivated on irradiated viral producer cells using infection medium consisting of Dulbecco modified Eagle medium, 15% fetal calf serum, 100 ng/mL IL-11, 50 ng/mL Fms-like tyrosine kinase 3 ligand, 100 ng/mL stem cell factor, 10 μg/mL ciprofloxacin (Serologicals), and 6 μg/mL polybrene (hexadimethrine bromide, Sigma-Aldrich).
Cell culture and characterization
Methylcellulose cultures.
Progenitor assays were plated and scored as described.13
CRU assay.
Stem cell quantification assays were performed essentially as described.16
Inverse PCR, comparative genomic hybridization, and spectral karyotyping.
These techniques were performed as described.17
Flow cytometry studies
Flow cytometric analyses were performed using an LSRII cytometer (BD Biosciences). Monoclonal antibodies were specific for the following: Sca-1 (D7), Mac1 (M1/70), Gr-1 (RB6–8C5), CD41 (MWReg30), CD19 (1D3), CD43 (S7), CD4 (H129.19), and CD8 (53–6.7) from BD Biosciences PharMingen; Kit (2B8), CD48 (HM48–1), CD34 (RAM34), CD71 (R17217), and CD45R (RA3–6B2) from eBioscience; and CD150 (TC15–12F12.2) and TER-119 from BioLegend. A mixture of monoclonal antibodies against CD45R, TER-119, and Gr-1 was used as a lineage marker (Lineage). Side population staining was performed as previously described.18 The peak excitation wavelength for oxidized 2-dichlorofluorescein was 488 nm and emission was 525 nm.
Collection of RNA FLA2/FLB1 cells
FLA2/FLB1 cells were thawed and then washed twice (175g for 10 minutes) in 2% fetal bovine serum (Invitrogen) in phosphate-buffered saline. A total of 2 × 105 cells/mouse was then injected into the tail vein of mice sublethally irradiated (700 cGy) C57BL/6 mice. Mice were killed approximately 4 weeks after injection, femur and tibia bones were removed, and bone marrow was flushed with 3 mL of phosphate-buffered saline. Bone marrow aspirates were centrifuged (400g for 2 minutes) and aspirated before being resuspended in 10 mL of Trizol (Invitrogen). RNA was extracted according to the manufacturer's protocols before being treated by RNase-free DNase (Ambion) for 30 minutes at 37°C to degrade any contaminating genomic DNA. After confirming RNA quality and integrity by Bioanalyzer (Agilent Technologies), rRNA was then specifically depleted from samples using the RiboMinus kit (Invitrogen) and polyadenylated transcripts were enriched using the Oligotex Direct mRNA kit (QIAGEN) according to their respective manufacturer's protocols.
Microarray analysis
Total RNA was reverse-transcribed (RT) using a Superscript double-stranded cDNA synthesis kit (Invitrogen). A total of 1 μg of either purified cDNA was labeled with Cy3- or Cy5-conjugated nonamers (TriLink BioTechnologies) using Klenow fragment (3′→5′ exo-) according to Nimblegen recommendations. Labeled samples were competitively hybridized on catalog mouse expression arrays (MM8_60mer expr, Roche-Nimblegen) before being washed and scanned according to Nimblegen recommendations. Signals for biologic replicates for each cell type were averaged, and RMA (Robust Multichip Average) normalized of raw expression data and gene expression calls was performed using Nimblescan, Version 2.5 software (Roche-Nimblegen). Raw and processed microarray data were submitted to ArrayExpress under accession number E-TABM-947.
SOLiD sequencing and sequence analysis
cDNA was sequenced according to the manufacturer's protocols for the SOLiD Total RNA-Seq kit for whole transcriptome (AB Life Sciences). Briefly, 10 μg of total RNA from each tell type was depleted of rRNA and enriched for PolyA transcripts as for microrarray work above, and 1 μg of this RNA was then fragmented using RNase III. Ligation of the adaptor mix A and reverse transcription were performed following the manufacturer's protocol. Libraries were size selected for fragments between 150 and 300 bp, amplified for 15 cycles of polymerase chain reaction (PCR), and purified using PureLink PCR micro kit (Invitrogen). Library concentrations were determined by quantitative PCR using a standard curve of template at known concentrations (DH10B), and approximately 0.25 ng of the FLA2/FLB1 libraries was used for each full emulsion PCR (emPCR) reaction (4 emPCR/sample). Approximately 300 millions of beads were deposited on a full slide and sequenced using the Opti Fragment Library Sequencing kit-Master Mix 50 on a SOLiD machine (Version 3).
Results
Hoxa9 + Meis1 generated leukemias with different L-HSC frequencies
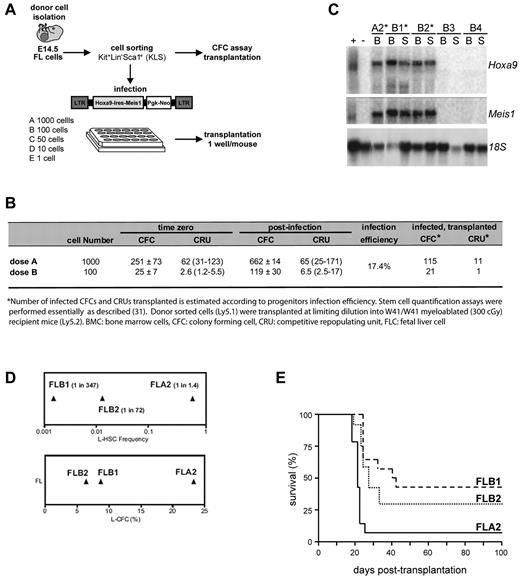
A subpopulation (Kit+Lin−Sca1+ [KLS]) highly enriched for stem cells and multipotent progenitors was isolated from FL and evaluated for frequency of colony-forming cells (CFCs) and competitive repopulation units (CRU). A fraction of these cells were distributed at various doses (Figure 1A) in 24-well plates coated with viral producer cells transfected with the murine stem cell virus Hoxa9-ires-Meis1-pgk-Neo retroviral plasmid, previously documented to provide a full oncogenic complement to primitive bone marrow cells.10 After infection, the contents of each well were either transferred into methylcellulose culture to assess expansion of progenitors and gene transfer efficiency, or directly transplanted into sublethally irradiated recipients to evaluate the stem cell content (Figure 1A). The higher cell dose transplanted (1000 KLS start cells, corresponding to 115 infected CFCs and 21 infected CRUs), as well as lower quantities of infected cells (100 KLS start cells, corresponding to 21 infected CFCs and one infected CRU) resulted in typical Hoxa9 + Meis1-induced myeloid leukemia occurring within the previously documented time frame19 (Figure 1B; and data not shown). An intact Hoxa9 + Meis1 provirus was observed in all leukemias analyzed, and both oncogenes were expressed at similar levels (Figure 1C). Of importance, leukemias did not occur in mice reconstituted with cell doses containing only transduced CFCs, and leukemia was only observed in mice that received cell fractions containing at least one stem cell (data not shown). These results suggest that Hoxa9 + Meis1 transformed only a subset of primitive hematopoietic cells.
Generation of a stem cell leukemia (FLA2). Overview of the experimental strategy. (A) E14.5 FL cells were isolated from C57BL/6:Pep3b (Ly5.1) mice and sorted for KLS cells. A fraction of sorted cells was plated at limiting dilution in 24-well plates containing murine stem cell virus Hoxa9-ires-Meis1a-pgk-Neo retroviral producers. Transduced cells were transplanted into congenic mice at a dose of one well per mouse. (B) Summary of CFC and transplanted CRU infection efficiency. Numbers of infected CFCs and CRUs were estimated according to progenitor infection percentage. (C) Northern blot analysis of Hoxa9 and Meis1 mRNA levels in leukemia (*) and healthy mice (B3 and B4) with 18S RNA levels were used as loading control. + indicates GP + E86 viral producers of retrovirus; and −, noninfected GP + E86 cells. (D) Leukemia colony-forming cell (L-CFC; bottom panel) and L-HSC frequencies (top panel) determined in primary recipients of FLA2, FLB2, and FLB1 cultures as measured by limiting dilution analyses. (E) Kaplan-Meier-like plot comparing survivals of secondary mice transplanted at limiting dilutions (ie, one L-HSC per recipient, corresponding to 1 cell for FLA2 and several hundred for FLB1 and FLB2) where the survival curves for FLA2 and FLB1 are significantly different (P = .0001).
Generation of a stem cell leukemia (FLA2). Overview of the experimental strategy. (A) E14.5 FL cells were isolated from C57BL/6:Pep3b (Ly5.1) mice and sorted for KLS cells. A fraction of sorted cells was plated at limiting dilution in 24-well plates containing murine stem cell virus Hoxa9-ires-Meis1a-pgk-Neo retroviral producers. Transduced cells were transplanted into congenic mice at a dose of one well per mouse. (B) Summary of CFC and transplanted CRU infection efficiency. Numbers of infected CFCs and CRUs were estimated according to progenitor infection percentage. (C) Northern blot analysis of Hoxa9 and Meis1 mRNA levels in leukemia (*) and healthy mice (B3 and B4) with 18S RNA levels were used as loading control. + indicates GP + E86 viral producers of retrovirus; and −, noninfected GP + E86 cells. (D) Leukemia colony-forming cell (L-CFC; bottom panel) and L-HSC frequencies (top panel) determined in primary recipients of FLA2, FLB2, and FLB1 cultures as measured by limiting dilution analyses. (E) Kaplan-Meier-like plot comparing survivals of secondary mice transplanted at limiting dilutions (ie, one L-HSC per recipient, corresponding to 1 cell for FLA2 and several hundred for FLB1 and FLB2) where the survival curves for FLA2 and FLB1 are significantly different (P = .0001).
To directly assess the L-HSC compartment, limiting dilution assays were performed in secondary mice, which showed that FL-derived leukemias have wide-ranging L-HSC frequencies, from 1 per 1.4 cells (FLA2 leukemia) to 1 per 347 (FLB1 leukemia) (Figure 1D top panel). Importantly, these L-HSC frequencies remained stable during successive transplantations and on multiple freeze/thaw procedures. Of note, leukemia aggressiveness also correlated with L-HSC frequency, and the shortest time from the injection of one L-HSC to clinical AML was obtained with FLA2 leukemia (Figure 1E; and data not shown).
FLA2 cells are similar to other Hoxa9 + Meis1-induced leukemias
A unique Hoxa9 + Meis1 retroviral insertion site in FLA2 cells was mapped to chromosome 2 by inverse PCR (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). However quantitative RT-PCR analysis revealed that none of the 20 genes within a 2-Mb distance from the integrated provirus was deregulated in FLA2 cells (data not shown). G banding, spectral karyotyping, and comparative genomic hybridization analyses confirmed that the specific biologic property of FLA2 cells was not consequent to coarse chromosomal abnormalities (supplemental Figure 2). Morphologic analysis, including dissemination characteristics (eg, spleen infiltration), also confirmed the high similarity between all generated leukemias (supplemental Figure 3A; and data not shown). Despite a striking difference in L-HSC frequency, cell surface markers revealed that FLA2 cells were similar to all other leukemias generated within the same experiment, each of which exhibited a homogeneous myeloid cell surface phenotype (Kit+Sca1−CD150−CD48+CD34+CD71+Mac1+GR1+; supplemental Figure 3B). FLA2 and FLB1 leukemias did not reveal any significant difference in the cell cycle phase distribution (data not shown); however, we did find that FLA2 leukemia contained a slightly elevated proportion of side population cells (supplemental Figure 3C), a fraction known in normal bone marrow to be enriched for cells with long-term reconstitution ability.20
Generation of transcriptome data for leukemic cell types by RNA-seq and microarrays
To analyze differences in the transcriptome content of the 2 cell types, RNA was collected and analyzed from FLA2 and FLB1 mice. Cells of each type were grafted into syngeneic mice, which were killed after 4 weeks when overgrowth of leukemic cells in bone marrow was close to 90% (verified by cytospins, data not shown). RNA from each biologic replicate (2 mice for each cell type) was collected and split for analysis by RNA-seq and by expression microarray. Sequencing of rRNA depleted, polyA+ enriched cDNA on the AB SOLiD, Version 3.0 machine generated 370 and 346 million reads for FLA2 and FLB1 replicates, respectively, of which approximately 60% could be mapped in both cases (Table 1). Similar to other studies, we have found that 1% to 3% of mapped reads span exon-exon junctions.21 In looking at the position of the mapped reads, we observed good specificity for the majority of reads to fall within annotated gene features, followed by introns (probably the result of isolation of unspliced mRNA) and intergenic regions (supplemental Figure 4). It is also clear that some of the signal in the last 2 classes probably comes from novel transcribed elements. Based on the discovery plot generated from our FLA2 (supplemental Figure 5), the depth of our sequencing is sufficiently deep enough to have discovered any directed transcription occurring.
RNA-seq statistics
| Statistic . | FLA2 replicate 1 . | FLA2 replicate 2 . | FLB1 replicate 1 . | FLB1 replicate 2 . |
|---|---|---|---|---|
| Total raw reads | 88312565 | 282300223 | 166168360 | 179984287 |
| Mapped reads | 51317865 | 194814974 | 99469306 | 122342813 |
| Reads mapped to splice junctions | 536577 | 6625312 | 1491603 | 4140951 |
| % mapped to junction | 1.05 | 3.40 | 1.50 | 3.38 |
| Aggregate total raw reads | 370612788 | 346152647 | ||
| Aggregate mapped reads | 246132839 | 221812119 | ||
| Aggregate sequence (Gbp) | 10.299 | 9.193 | ||
| Statistic . | FLA2 replicate 1 . | FLA2 replicate 2 . | FLB1 replicate 1 . | FLB1 replicate 2 . |
|---|---|---|---|---|
| Total raw reads | 88312565 | 282300223 | 166168360 | 179984287 |
| Mapped reads | 51317865 | 194814974 | 99469306 | 122342813 |
| Reads mapped to splice junctions | 536577 | 6625312 | 1491603 | 4140951 |
| % mapped to junction | 1.05 | 3.40 | 1.50 | 3.38 |
| Aggregate total raw reads | 370612788 | 346152647 | ||
| Aggregate mapped reads | 246132839 | 221812119 | ||
| Aggregate sequence (Gbp) | 10.299 | 9.193 | ||
Biologic replicates of leukemic cells show good concordance irrespective of assay method
To compare RNAseq data to microarray data, gene models were constructed by downloading the all_mrna table from University of California Santa Cruz (UCSC),22 which contains all non-EST (expressed sequence tag) mRNAs, which have been sequenced. These mRNAs were compared with themselves to find all splicing variants within a given loci. Microarray and RNA-seq data were assigned to specific genes, and the concordance of signals between biologic replicates was analyzed (Figure 2). All RNA samples showed highly consistent, and significant, correlation coefficients regardless of whether they were sequenced or hybridized on microarrays. Although the FLA2 samples did show somewhat lower correlations, the fact that this was consistently observed suggests that there was a slight biologic difference between the 2 FLA2 replicates. Given the consistency of the results, the microarray scores were averaged between replicates and the RNA-seq data pooled to simplify downstream analysis.
Comparison of biologic replicates by platform. Expression scores of genes for each biologic replicate of RNA-seq (top row) or microarray (bottom row) are shown plotted on log10 scales. Linear models (red line) and concordance scores with P values were calculated using the lm and concordance.lm R functions and demonstrate good reproducibility regardless of assay platform used.
Comparison of biologic replicates by platform. Expression scores of genes for each biologic replicate of RNA-seq (top row) or microarray (bottom row) are shown plotted on log10 scales. Linear models (red line) and concordance scores with P values were calculated using the lm and concordance.lm R functions and demonstrate good reproducibility regardless of assay platform used.
RNA-seq allows more sensitive monitoring of transcriptional changes in cells than microarrays
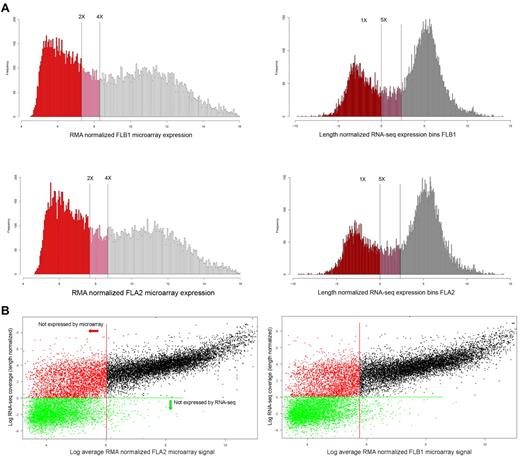
Expression histograms were generated using all genes in common to both platforms, which could be uniquely identified by a single Entrez gene ID to classify genes according to their distribution within all signals (Figure 3). As expected, the dynamic range of RNA-seq was far greater than that of the microarrays, with a greater than 5000-fold difference in their respective range of measurements.
Expression histograms and platform comparisons. (A) Expression histograms based on microarray data (left column) and RNA-seq results (right column). The log2 value of the length normalized expression levels of histogram bins is plotted along the x-axis, whereas the number of genes falling into each bins is plotted along the y-axis. Thresholds for “no expression” or “marginal expression” are shown as vertical lines at either 2-fold and 4-fold above GC control probes signals (microarray) or 1-fold and 5-fold length normalized coverage (RNA-seq), respectively. Genes classed as not expressed are colored in red, marginally expressed genes in pink, and expressed as empty bars. (B) Individual gene expression levels (log2) are shown plotted by RNA-seq (y-axis) and by microarray (x-axis) values for either FLA2 (top panel) or FLB1 (bottom panel). Genes below marginal expression levels for microarray (2-fold GC controls, Figure 5) are shown left of the red vertical line. Genes below marginal expression levels for RNA-seq (1-fold LN coverage in panel A) are shown below the green horizontal line. The increased sensitivity of RNA-seq expression values is apparent in the 2 populations of genes visible left of the red vertical line, which all represent nonexpressed genes by microarray.
Expression histograms and platform comparisons. (A) Expression histograms based on microarray data (left column) and RNA-seq results (right column). The log2 value of the length normalized expression levels of histogram bins is plotted along the x-axis, whereas the number of genes falling into each bins is plotted along the y-axis. Thresholds for “no expression” or “marginal expression” are shown as vertical lines at either 2-fold and 4-fold above GC control probes signals (microarray) or 1-fold and 5-fold length normalized coverage (RNA-seq), respectively. Genes classed as not expressed are colored in red, marginally expressed genes in pink, and expressed as empty bars. (B) Individual gene expression levels (log2) are shown plotted by RNA-seq (y-axis) and by microarray (x-axis) values for either FLA2 (top panel) or FLB1 (bottom panel). Genes below marginal expression levels for microarray (2-fold GC controls, Figure 5) are shown left of the red vertical line. Genes below marginal expression levels for RNA-seq (1-fold LN coverage in panel A) are shown below the green horizontal line. The increased sensitivity of RNA-seq expression values is apparent in the 2 populations of genes visible left of the red vertical line, which all represent nonexpressed genes by microarray.
Given the arbitrary nature of thresholds chosen for calling a gene “expressed,” we elected to set 2 thresholds, one for nonexpression and one for marginal expression, as has been done in previous studies.23 For the microarray data, 6131 probes corresponding to random GC (guanine-cytosine content) content controls were used to assess background signals, as these probes should not hybridize to anything within the samples. The threshold for expression by microarray was set at twice the level of signal from the average of the GC control probes in each channel, whereas the limit for marginal expression was set as 4 times the average GC control signal. Using these thresholds, there were 8480 and 9031 genes expressed and 1646 and 1758 genes marginally expressed of the total of 17 262 genes (analyzed in common between microarray and RNA-seq experiments) in FLA2 and FLB1 cells, respectively.
For the RNA-seq equivalent, thresholds were set at 1-fold length normalized coverage, and 5-fold length normalized coverage for expression, based on the distribution of signals. At these thresholds, we found there were 12 115 and 11 866 transcripts expressed and 1724 and 1641 transcripts marginally expressed of the total 17 262. Thus, although the number of marginally expressed genes is similar with both platforms, the greater dynamic range of RNA-seq results in more genes being categorized as being expressed.
The increased sensitivity of RNA-seq is also evident in a scatterplot of signals of each gene from each platform (Figure 3). The threshold for expression for genes by either microarray (red) or RNA-seq (green) is shown by either vertical or horizontal lines, respectively. In both cell types, the pattern of signals is similar with a fairly linear range down to the expression threshold for microarrays. Beyond this point, although all genes are not called as expressed by microarray, 2 populations of genes are visible: those that are expressed by RNA-seq (red dots) and those that are not expressed by either microarray or RNA-seq (green dots).
FLA2/FLB1 cells have differentially expressed genes despite overexpressing the same 2 genes
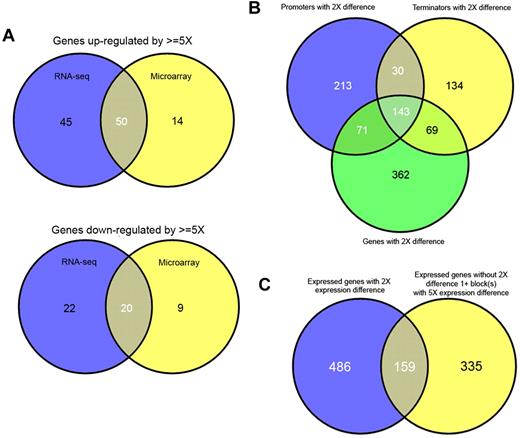
Unexpectedly, these 2 leukemias contained a large number of differentially expressed genes. At a threshold of 2-fold up- or down-regulation, 647 and 1072 genes are found to be differentially regulated, and above marginal expression levels, by RNA-seq and microarray, respectively (supplemental Table 1); however, many of these ratios are only marginally over 2-fold. Using a more conservative cutoff of 5-fold reduces the number of differentially expressed genes to 137 and 93, of which the top 50 by RNA-seq are shown (Table 2). Despite the absence of probes for several genes on the microarrays, or marginal expression of others, both methodologies showed good overlap. Therefore, when gene lists using conservative cut-offs are compared, it is evident that there is good correlation between both methods and that genes determined to be differentially regulated by microarray are largely a subset of those determined by RNA-seq (Figure 4A).
Genes up-regulated/down-regulated in FLA2/FLB1
| Transcript . | RNA-seq rank . | RNA-seq ratio (FLB1/FLA2) . | FLA2 coverage . | FLB1 coverage . | Microarray rank . | Microarray ratio (FLB1/FLA2) . | Gene name . | Description . |
|---|---|---|---|---|---|---|---|---|
| AK090126 | 1 | 72.64 | 27.5 | 1999.7 | 1 | 23.25 | Mela | Melanoma antigen |
| AF319945 | 2 | 52.17 | 0.2 | 11.1 | NOA | NA | Nespas | Neuroendocrine secretory protein antisense |
| AK083486 | 3 | 35.33 | 0.2 | 6.6 | NOA | NA | D2Ertd173e | DNA segment, Chr 2, ERATO Doi 173, expressed |
| BC100365 | 4 | 28.09 | 0.6 | 15.5 | 3 | 13.69 | 1700030C10Rik | RIKEN cDNA 1700030C10 gene |
| X59289 | 5 | 23.13 | 16.8 | 356.0 | NOA | NA | Xist | Inactive X specific transcripts |
| AK007645 | 6 | 21.21 | 0.4 | 8.5 | M/NE | NA | Gal3st1 | Galactose-3-O-sulfotransferase 1 |
| AK147315 | 7 | 20.04 | 20.8 | 417.5 | 4 | 11.90 | Fn1 | Fibronectin 1 |
| AK132299 | 8 | 17.65 | 0.5 | 9.5 | 11 | 8.00 | Susd5 | Sushi domain containing 5 |
| BC064108 | 9 | 16.92 | 0.5 | 8.9 | 69 | 3.00 | 5830444B04Rik | RIKEN cDNA 5830444B04 gene |
| BC023460 | 10 | 16.52 | 0.4 | 5.1 | NOA | NA | Fam89a | Family with sequence similarity 89, member A |
| AK166253 | 11 | 16.15 | 1.2 | 19.9 | 7 | 11.36 | Smpdl3b | Sphingomyelin phosphodiesterase, acid-like 3B |
| AK052580 | 12 | 12.64 | 3.1 | 37.5 | 4 | 11.76 | Serpinb10 | Serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10 |
| BC139010 | 13 | 12.61 | 9.4 | 118.5 | 2 | 24.39 | Stag3 | Stromal antigen 3 |
| AK219870 | 14 | 12.42 | 0.5 | 6.4 | NOA | NA | Rnf217 | Ring finger protein 217 |
| AK034871 | 15 | 11.75 | 7.7 | 90.6 | 13 | 7.14 | Vcan | Versican |
| AK146132 | 16 | 11.49 | 0.9 | 10.3 | M/NE | NA | Gzma | Granzyme A |
| AK040922 | 17 | 10.84 | 0.6 | 6.7 | 69 | 3.00 | 2010015L04Rik | RIKEN cDNA 2010015L04 gene |
| AK161020 | 18 | 10.45 | 1.2 | 12.4 | 6 | 11.49 | Ace | Angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| BC016111 | 19 | 9.29 | 1.7 | 15.3 | 5 | 11.62 | Grb10 | Growth factor receptor bound protein 10 |
| AF488553 | 20 | 8.59 | 0.9 | 7.9 | 28 | 5.02 | Dpep3 | Dipeptidase 3 |
| AF253057 | 21 | 8.43 | 10.2 | 85.8 | 26 | 5.31 | Klra2 | Killer cell lectin-like receptor, subfamily A, member 2 |
| AK008099 | 22 | 8.23 | 1.1 | 9.2 | M/NE | NA | Ms4a8a | Membrane-spanning 4-domains, subfamily A, member 8A |
| L04275 | 23 | 7.77 | 4.3 | 33.2 | 10 | 9.52 | Msr1 | Macrophage scavenger receptor 1 |
| AK132144 | 24 | 7.62 | 0.7 | 5.1 | 9 | 10.86 | Sec16b | SEC16 homolog B (S. cerevisiae) |
| AK157856 | 25 | 7.16 | 1.1 | 7.6 | M/NE | NA | Gzmb | Granzyme B |
| AK088637 | 1 | 31.03 | 75.7 | 2.4 | NOA | NA | B230118H07Rik | RIKEN cDNA B230118H07 gene |
| BC160356 | 2 | 24.37 | 408.8 | 16.6 | 1 | 43.31 | Olfm4 | Olfactomedin 4 |
| AK027937 | 3 | 21.57 | 17.9 | 0.8 | 31 | 10.46 | Mpp7 | Membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| S57123 | 4 | 19.95 | 16343.7 | 815.0 | 5 | 23.97 | S100a8 | S-100 calcium-binding protein A8 (calgranulin A) |
| AK143826 | 5 | 19.74 | 6043.3 | 306.1 | 29 | 10.68 | S100a9 | S-100 calcium-binding protein A9 (calgranulin B) |
| BC146444 | 6 | 19.22 | 54.7 | 2.8 | 14 | 16.41 | Gm5416 | Predicted gene 5416 |
| BC049566 | 7 | 19.19 | 95.1 | 5.0 | 4 | 24.46 | Fundc2 | FUN14 domain containing 2 |
| AY042192 | 8 | 19.17 | 27.9 | 1.5 | 8 | 21.87 | Mrgpra2b | MAS-related GPR, member A2B |
| BC064040 | 9 | 19.11 | 30.7 | 1.6 | 2 | 36.41 | Mrgpra2a | MAS-related GPR, member A2A |
| AK158005 | 10 | 18.37 | 6337.3 | 345.1 | 16 | 16.06 | Camp | Cathelicidin antimicrobial peptide |
| BC107220 | 11 | 17.31 | 692.9 | 40.0 | 20 | 13.97 | Fcnb | Ficolin B |
| BC008530 | 12 | 17.27 | 1004.7 | 58.2 | 18 | 14.97 | Ltf | Lactotransferrin |
| AK035520 | 13 | 16.69 | 6.5 | 0.4 | NOA | NA | Gm5101 | Predicted gene 5101 |
| AJ514933 | 14 | 16.10 | 149.9 | 9.3 | 3 | 24.65 | Retnlg | Resistin-like γ |
| BC147081 | 15 | 16.09 | 1971.4 | 122.5 | 12 | 17.60 | Gm5483 | Predicted gene 5483 |
| AK003352 | 16 | 15.79 | 242.9 | 15.4 | 50 | 6.53 | 1100001G20Rik | RIKEN cDNA 1100001G20 gene |
| AK141338 | 17 | 15.42 | 6.1 | 0.4 | M/NE | NA | Il28ra | Interleukin 28 receptor alpha |
| BC119403 | 18 | 15.11 | 7881.0 | 521.5 | 48 | 6.79 | Ngp | Neutrophilic granule protein |
| BC079631 | 19 | 14.94 | 32.6 | 2.2 | 72 | 4.55 | Septin5 | Septin 5 |
| M92418 | 20 | 14.93 | 408.5 | 27.4 | 10 | 19.96 | Stfa2 | Stefin A2 |
| AY163161 | 21 | 14.66 | 658.2 | 44.7 | 13 | 17.47 | Stfa2l1 | Stefin A2-like 1 |
| BC125396 | 22 | 14.33 | 323.5 | 22.6 | NOA | NA | 2010005H15Rik | RIKEN cDNA 2010005H15 gene |
| AF051367 | 23 | 14.22 | 179.0 | 12.5 | 24 | 11.79 | Itgb2l | Integrin-β2-like |
| BC100530 | 24 | 14.06 | 1542.0 | 109.7 | 1897 | 1.77 | BC100530 | cDNA sequence BC100530 |
| AK008704 | 25 | 14.03 | 43.6 | 3.1 | 7 | 22.13 | Pttg1 | Pituitary tumor-transforming gene 1 |
| Transcript . | RNA-seq rank . | RNA-seq ratio (FLB1/FLA2) . | FLA2 coverage . | FLB1 coverage . | Microarray rank . | Microarray ratio (FLB1/FLA2) . | Gene name . | Description . |
|---|---|---|---|---|---|---|---|---|
| AK090126 | 1 | 72.64 | 27.5 | 1999.7 | 1 | 23.25 | Mela | Melanoma antigen |
| AF319945 | 2 | 52.17 | 0.2 | 11.1 | NOA | NA | Nespas | Neuroendocrine secretory protein antisense |
| AK083486 | 3 | 35.33 | 0.2 | 6.6 | NOA | NA | D2Ertd173e | DNA segment, Chr 2, ERATO Doi 173, expressed |
| BC100365 | 4 | 28.09 | 0.6 | 15.5 | 3 | 13.69 | 1700030C10Rik | RIKEN cDNA 1700030C10 gene |
| X59289 | 5 | 23.13 | 16.8 | 356.0 | NOA | NA | Xist | Inactive X specific transcripts |
| AK007645 | 6 | 21.21 | 0.4 | 8.5 | M/NE | NA | Gal3st1 | Galactose-3-O-sulfotransferase 1 |
| AK147315 | 7 | 20.04 | 20.8 | 417.5 | 4 | 11.90 | Fn1 | Fibronectin 1 |
| AK132299 | 8 | 17.65 | 0.5 | 9.5 | 11 | 8.00 | Susd5 | Sushi domain containing 5 |
| BC064108 | 9 | 16.92 | 0.5 | 8.9 | 69 | 3.00 | 5830444B04Rik | RIKEN cDNA 5830444B04 gene |
| BC023460 | 10 | 16.52 | 0.4 | 5.1 | NOA | NA | Fam89a | Family with sequence similarity 89, member A |
| AK166253 | 11 | 16.15 | 1.2 | 19.9 | 7 | 11.36 | Smpdl3b | Sphingomyelin phosphodiesterase, acid-like 3B |
| AK052580 | 12 | 12.64 | 3.1 | 37.5 | 4 | 11.76 | Serpinb10 | Serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10 |
| BC139010 | 13 | 12.61 | 9.4 | 118.5 | 2 | 24.39 | Stag3 | Stromal antigen 3 |
| AK219870 | 14 | 12.42 | 0.5 | 6.4 | NOA | NA | Rnf217 | Ring finger protein 217 |
| AK034871 | 15 | 11.75 | 7.7 | 90.6 | 13 | 7.14 | Vcan | Versican |
| AK146132 | 16 | 11.49 | 0.9 | 10.3 | M/NE | NA | Gzma | Granzyme A |
| AK040922 | 17 | 10.84 | 0.6 | 6.7 | 69 | 3.00 | 2010015L04Rik | RIKEN cDNA 2010015L04 gene |
| AK161020 | 18 | 10.45 | 1.2 | 12.4 | 6 | 11.49 | Ace | Angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| BC016111 | 19 | 9.29 | 1.7 | 15.3 | 5 | 11.62 | Grb10 | Growth factor receptor bound protein 10 |
| AF488553 | 20 | 8.59 | 0.9 | 7.9 | 28 | 5.02 | Dpep3 | Dipeptidase 3 |
| AF253057 | 21 | 8.43 | 10.2 | 85.8 | 26 | 5.31 | Klra2 | Killer cell lectin-like receptor, subfamily A, member 2 |
| AK008099 | 22 | 8.23 | 1.1 | 9.2 | M/NE | NA | Ms4a8a | Membrane-spanning 4-domains, subfamily A, member 8A |
| L04275 | 23 | 7.77 | 4.3 | 33.2 | 10 | 9.52 | Msr1 | Macrophage scavenger receptor 1 |
| AK132144 | 24 | 7.62 | 0.7 | 5.1 | 9 | 10.86 | Sec16b | SEC16 homolog B (S. cerevisiae) |
| AK157856 | 25 | 7.16 | 1.1 | 7.6 | M/NE | NA | Gzmb | Granzyme B |
| AK088637 | 1 | 31.03 | 75.7 | 2.4 | NOA | NA | B230118H07Rik | RIKEN cDNA B230118H07 gene |
| BC160356 | 2 | 24.37 | 408.8 | 16.6 | 1 | 43.31 | Olfm4 | Olfactomedin 4 |
| AK027937 | 3 | 21.57 | 17.9 | 0.8 | 31 | 10.46 | Mpp7 | Membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| S57123 | 4 | 19.95 | 16343.7 | 815.0 | 5 | 23.97 | S100a8 | S-100 calcium-binding protein A8 (calgranulin A) |
| AK143826 | 5 | 19.74 | 6043.3 | 306.1 | 29 | 10.68 | S100a9 | S-100 calcium-binding protein A9 (calgranulin B) |
| BC146444 | 6 | 19.22 | 54.7 | 2.8 | 14 | 16.41 | Gm5416 | Predicted gene 5416 |
| BC049566 | 7 | 19.19 | 95.1 | 5.0 | 4 | 24.46 | Fundc2 | FUN14 domain containing 2 |
| AY042192 | 8 | 19.17 | 27.9 | 1.5 | 8 | 21.87 | Mrgpra2b | MAS-related GPR, member A2B |
| BC064040 | 9 | 19.11 | 30.7 | 1.6 | 2 | 36.41 | Mrgpra2a | MAS-related GPR, member A2A |
| AK158005 | 10 | 18.37 | 6337.3 | 345.1 | 16 | 16.06 | Camp | Cathelicidin antimicrobial peptide |
| BC107220 | 11 | 17.31 | 692.9 | 40.0 | 20 | 13.97 | Fcnb | Ficolin B |
| BC008530 | 12 | 17.27 | 1004.7 | 58.2 | 18 | 14.97 | Ltf | Lactotransferrin |
| AK035520 | 13 | 16.69 | 6.5 | 0.4 | NOA | NA | Gm5101 | Predicted gene 5101 |
| AJ514933 | 14 | 16.10 | 149.9 | 9.3 | 3 | 24.65 | Retnlg | Resistin-like γ |
| BC147081 | 15 | 16.09 | 1971.4 | 122.5 | 12 | 17.60 | Gm5483 | Predicted gene 5483 |
| AK003352 | 16 | 15.79 | 242.9 | 15.4 | 50 | 6.53 | 1100001G20Rik | RIKEN cDNA 1100001G20 gene |
| AK141338 | 17 | 15.42 | 6.1 | 0.4 | M/NE | NA | Il28ra | Interleukin 28 receptor alpha |
| BC119403 | 18 | 15.11 | 7881.0 | 521.5 | 48 | 6.79 | Ngp | Neutrophilic granule protein |
| BC079631 | 19 | 14.94 | 32.6 | 2.2 | 72 | 4.55 | Septin5 | Septin 5 |
| M92418 | 20 | 14.93 | 408.5 | 27.4 | 10 | 19.96 | Stfa2 | Stefin A2 |
| AY163161 | 21 | 14.66 | 658.2 | 44.7 | 13 | 17.47 | Stfa2l1 | Stefin A2-like 1 |
| BC125396 | 22 | 14.33 | 323.5 | 22.6 | NOA | NA | 2010005H15Rik | RIKEN cDNA 2010005H15 gene |
| AF051367 | 23 | 14.22 | 179.0 | 12.5 | 24 | 11.79 | Itgb2l | Integrin-β2-like |
| BC100530 | 24 | 14.06 | 1542.0 | 109.7 | 1897 | 1.77 | BC100530 | cDNA sequence BC100530 |
| AK008704 | 25 | 14.03 | 43.6 | 3.1 | 7 | 22.13 | Pttg1 | Pituitary tumor-transforming gene 1 |
Comparison of structural variations and differentially expression genes within transcriptomes. The overlap between genes determined to be up-regulated (top) or down-regulated (bottom) by RNA-seq (blue) or microarray (yellow) is shown by Venn diagrams (A). Genes up-regulated/down-regulated by microarray generally represent a subset of genes up-regulated by RNA-seq. The overlap between promoters, terminators, and genes, which show a differential usage/expression of more than 2-fold, are shown (B). The overlap between genes with differential expression (> 2-fold) and blocks (exons or parts of exons), which show differential expression (> 5-fold) but are contained with genes, which do not show differential expression are shown (C).
Comparison of structural variations and differentially expression genes within transcriptomes. The overlap between genes determined to be up-regulated (top) or down-regulated (bottom) by RNA-seq (blue) or microarray (yellow) is shown by Venn diagrams (A). Genes up-regulated/down-regulated by microarray generally represent a subset of genes up-regulated by RNA-seq. The overlap between promoters, terminators, and genes, which show a differential usage/expression of more than 2-fold, are shown (B). The overlap between genes with differential expression (> 2-fold) and blocks (exons or parts of exons), which show differential expression (> 5-fold) but are contained with genes, which do not show differential expression are shown (C).
Specific analysis of mouse miRNAs, obtained from miRBase,24 showed that, of the 567 annotated elements, remarkably few are highly expressed, with the top 15 miRNAs (2.6%) by expression level accounting for more than 95% of all transcription from miRNAs (supplemental Table 2). Of the miRNAs that are expressed at a level probably biologically significant, only 2 (mmu-mir-2140 and mmu-mir-2133–1) exhibit differential expression between cell types (fla2:flb1 of 0.47 and 3.17, respectively). Given the lack of experimental validation of targets of these miRNAs and the multiplicity of computationally predicted targets, it is not possible to clearly identify a mechanism by which these miRNAs would contribute to the cellular behavior differences; however, this possibility cannot be ruled out without further investigations.
Differentially expressed genes have a biased distribution
To characterize the genes showing differential regulation, we analyzed the Gene Ontology (GO) annotation25 associated with the 310 genes exhibiting a greater than 3-fold difference in expression by RNA-seq. Analysis using the functional annotation clustering feature of DAVID26 showed that the highest scoring annotation cluster is enriched in genes associated with GO terms for immune system development, hemopoietic or lymphoid organ development, hemopoiesis, and myeloid cell differentiation (data not shown). The enrichment in GO terms associated with blood development and differentiation suggest that the differential expression of these genes is consistent with the differences in self-renewal capacity exhibited by the cell types.
As an alternative method to ascertain biases in GO term annotation with respect to differentially expressed genes, the GO terms for a wide variety of cellular processes were selected from AMIGO. These classes were then used to determine the percentage of genes expressed in each class (by RNA-seq) and then, of these, the number that were differentially expressed (Table 3; supplemental Table 3). Interestingly, whereas most processes had 50% to 80% of their associated genes being expressed and 2% to 6% of the genes differentially expressed, G-protein-coupled receptor activity was a notable exception where only approximately 5% of genes were expressed but approximately 22% were differentially expressed (Table 3). This effect is unlikely to simply be the result of size differences between classes given that the G-protein-coupled receptor and developmental process classes have approximately similar numbers of members but a percentage of expressed and differentially expressed genes that differs significantly between the 2.
Differential expression by GO category
| GO category . | Total in class . | Expressed in class, no. . | Expressed in class, % . | Differentially expressed, no. . | Differentially expressed, % . |
|---|---|---|---|---|---|
| Transcriptional activation | 288 | 136 | 47.22 | 7 | 5.15 |
| Transcriptional repression | 227 | 113 | 49.78 | 7 | 6.19 |
| Translational regulation | 82 | 64 | 78.05 | 4 | 6.25 |
| Chomosome organization | 64 | 49 | 76.56 | 2 | 4.08 |
| Kinase activity | 382 | 261 | 68.32 | 11 | 4.21 |
| GPCR activity | 1258 | 66 | 5.25 | 15 | 22.73 |
| Nuclear export activity | 24 | 21 | 87.50 | 1 | 4.76 |
| Developmental process | 1247 | 554 | 44.43 | 52 | 9.39 |
| Chromatin organization | 308 | 265 | 86.04 | 7 | 2.64 |
| GO category . | Total in class . | Expressed in class, no. . | Expressed in class, % . | Differentially expressed, no. . | Differentially expressed, % . |
|---|---|---|---|---|---|
| Transcriptional activation | 288 | 136 | 47.22 | 7 | 5.15 |
| Transcriptional repression | 227 | 113 | 49.78 | 7 | 6.19 |
| Translational regulation | 82 | 64 | 78.05 | 4 | 6.25 |
| Chomosome organization | 64 | 49 | 76.56 | 2 | 4.08 |
| Kinase activity | 382 | 261 | 68.32 | 11 | 4.21 |
| GPCR activity | 1258 | 66 | 5.25 | 15 | 22.73 |
| Nuclear export activity | 24 | 21 | 87.50 | 1 | 4.76 |
| Developmental process | 1247 | 554 | 44.43 | 52 | 9.39 |
| Chromatin organization | 308 | 265 | 86.04 | 7 | 2.64 |
GPCR indicates G-protein-coupled receptor.
The transcriptomes of FLA2/FLB1 exhibit numerous structural differences
Given the potential to analyze transcript structure using RNA-seq data,27 we undertook to characterize the differential use of alternative promoters, terminators, and exons within the transciptomes of FLA2/FLB1. To begin with, we computationally defined all single and multiple promoters and terminators based on all mRNAs downloaded from UCSC and measured the usage of these elements based on coverage by cell type (Tables 4,5). Interestingly, a strong bias toward the use of multiple promoters and terminators, where they exist, was observed, with approximately two-thirds of all being used in preference to a single promoter or terminator.
Statistics of alternative promoter usage
| . | Single promoters FLA2 . | Single promoters FLB1 . | Multiple promoters FLA2 . | Multiple promoters FLB1 . |
|---|---|---|---|---|
| Determined | 15699 | 15699 | 6046 | 6046 |
| Nonexpressed | 9975 | 9959 | 2473 | 2490 |
| Expressed from single promoters | 5724 | 5740 | 1049 | 1026 |
| Expressed from multiple promoters | NA | NA | 2524 | 2530 |
| . | Single promoters FLA2 . | Single promoters FLB1 . | Multiple promoters FLA2 . | Multiple promoters FLB1 . |
|---|---|---|---|---|
| Determined | 15699 | 15699 | 6046 | 6046 |
| Nonexpressed | 9975 | 9959 | 2473 | 2490 |
| Expressed from single promoters | 5724 | 5740 | 1049 | 1026 |
| Expressed from multiple promoters | NA | NA | 2524 | 2530 |
NA indicates not applicable.
Statistics of alternative terminator usage
| . | Single terminators FLA2 . | Single terminators FLB1 . | Multiple terminators FLA2 . | Multiple terminators FLB1 . |
|---|---|---|---|---|
| Determined | 15947 | 15947 | 5798 | 5798 |
| Nonexpressed | 10055 | 10136 | 2314 | 2352 |
| Terminated at single sites | 5892 | 5811 | 979 | 914 |
| Terminated at multiple sites | NA | NA | 2505 | 2532 |
| . | Single terminators FLA2 . | Single terminators FLB1 . | Multiple terminators FLA2 . | Multiple terminators FLB1 . |
|---|---|---|---|---|
| Determined | 15947 | 15947 | 5798 | 5798 |
| Nonexpressed | 10055 | 10136 | 2314 | 2352 |
| Terminated at single sites | 5892 | 5811 | 979 | 914 |
| Terminated at multiple sites | NA | NA | 2505 | 2532 |
NA indicates not applicable.
When we compared all promoters and terminators that had a more than 2-fold differential expression/usage, we found, as expected, that a majority of these features belonged to genes that also had an overall 2-fold difference (Figure 4B). For the 213 promoters and 134 terminators that did not overlap, we could see examples of alternative usage by cell type (supplemental Figure 6A). However, it is also probable that some of these nonoverlapping promoters and terminators result from slight differences in coverage of elements close to the threshold.
We also examined all exons for evidence of differential usage and again could find evidence for exons that showed differential usage when the overall genes did not. A large majority (67%) of exons that exhibited a 5-fold or greater differential usage were located within genes that did not have more than an overall 2-fold differential expression (Figure 4C). Although the same caveat regarding expression differences at or near thresholds applies, the higher threshold for exon usage mitigates this to a large degree. Through manual examination, we can again find clear examples of differential splicing without differential expression (supplemental Figure 6B-C).
Identification of SNV differences in cell types
The nucleotide resolution of the RNA-seq data allowed us to identify single nucleotide variations (SNVs) in either cell line with respect to the public reference sequence. To restrict our analysis to positions with sufficient sequence coverage, we required at least 20 reads covering a position in aggregate between the 2 biologic replicates before including it. Comparing commercial inbred mouse strains with the related reference sequence, we could find 1825 SNVs (supplemental Table 4) within mRNAs, which were identical between our cell types but differed synonymously from the reference sequence. In addition to these changes, we could detect 470 nonsynonymous SNVs in either cell relative to the reference sequence. These totals would seem high given the relative similarity of the cells, and their derivation ultimately from the same mouse strain despite their extended passaging in vivo; and so, similar to other deep sequencing studies,28,29 a large majority of these changes probably represent sequencing errors. To apply more restrictive selection criteria, when only consistent calls between biologic replicates are analyzed (whether heterozygous or homozygous) are used, there are 74 nonsynonymous changes identified. Interestingly, these changes include a mutation in the Meis1 gene, where the C at genomic position 18911310 in the reference is consistently called a C in FLA2 cells (413 reads), whereas it is consistently called a T in FLB1 cells (781 reads). This SNV is predicted to change an aspartic acid to an asparagine in the HR1 region previously identified to be important for interaction with Pbx1.30 A small percentage of the changes were also predicted to induce premature stop codons in the canonical protein sequence. We attempted to validate 10 of the predicted nonsynonymous changes, through sequencing-selected PCR-amplified regions from the cDNA. We were able to validate 2 of the changes, including the change in Meis1, which although somewhat low, agrees with the validation rate in similar studies.28,29
The genomes of FLA2/FLB1 contain numerous unannotated transcribed elements
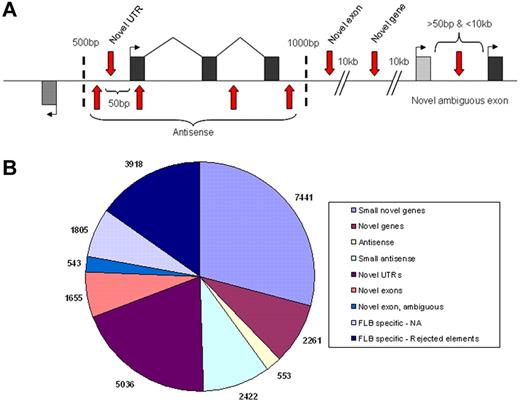
Although our analysis focused on annotated features derived from cloned mRNAs, we also sought to identify novel transcribed regions using our RNA-seq data. Using a 50-bp sliding window approach, we defined novel elements as starting when they had a minimum coverage level of 3-fold within the window and ending once the coverage dropped below this limit. Novel elements identified with this approach were required to have a minimum size of 75 bp and a maximum overlap of 25% with annotated repetitive elements to be included. To further reduce the number of false positive elements in the list, we then applied a highly conservative minimum expression level of 16-fold (a log2 value of 4 on Figure 3) for all elements (supplemental Table 5) and sought to classify all remaining elements into various categories as depicted (Figure 5A).
Classification of novel transcribed elements discovered. Exons within model genes are represented as black or gray exons, with the first exons marked by a bent arrow indicating the direction of transcription and vertical red arrows representing the various possible locations of novel transcribed elements (A). Locations relative to the model gene (shown in base pairs) represent the distant limits used for the various categories. The number of novel elements found within each category are shown (B) with small novel genes and small antisense being separated from their parent categories by a size limit of 150 bp.
Classification of novel transcribed elements discovered. Exons within model genes are represented as black or gray exons, with the first exons marked by a bent arrow indicating the direction of transcription and vertical red arrows representing the various possible locations of novel transcribed elements (A). Locations relative to the model gene (shown in base pairs) represent the distant limits used for the various categories. The number of novel elements found within each category are shown (B) with small novel genes and small antisense being separated from their parent categories by a size limit of 150 bp.
Most novel elements identified in FLA2 cells (supplemental Figure 7B) were relatively small in size, with a size distribution centered around approximately 120 bp in size (supplemental Figure 8). As a result, we partitioned the 2 largest classes of elements unrelated to known genes (antisense and novel genes) into 2 size groups, those above or below 150 bp in size, to avoid potentially mixing 2 distinct populations of elements. As demonstrated by the summary of elements (Table 6), with the exception of novel untranslated regions (which represent extensions of known mRNAs), more than 75% of novel elements in FLA2 cells are found within the same class in FLB1 cells, suggesting that these elements are genuine unannotated genes/exons. To find additional evidence that these novel elements are the result of directed transcription, we looked to determine whether they overlapped with regions of genome, which show strong levels of evolutionary conservation in the PhastCons Conserved Elements31 track of UCSC. When we examined only the novel elements that showed any degree of overlap with PhastCons elements, the average percentage of base pairs covered by PhastCons regions in these elements is very high (∼ 50%-67%). In addition to this evidence, in many (though not all) cases, we were able to identify strong EST support for transcription in the regions as well as Ensembl gene models (supplemental Figure 4).
Summary of statistics for novel elements
| . | No. found FLA2 . | Average LN coverage FLA2 (median) . | Found in same class FLB1, no. . | Found in same class FLB1, % . | Average PhastCoverage . | PhastCoverage excluding nonoverlapping, no. . | PhastCoverage excluding nonoverlapping, % . |
|---|---|---|---|---|---|---|---|
| Novel gene | 2261 | 270 (34) | 1924 | 85.1 | 33.7 | 968 | 67.0 |
| Small novel gene | 7441 | 79 (28) | 5449 | 73.2 | 6 | 576 | 56.9 |
| Antisense | 553 | 185 (33) | 458 | 82.8 | 30 | 221 | 61.7 |
| Small antisense | 2422 | 84 (28) | 1819 | 75.1 | 12.2 | 403 | 54.7 |
| Novel UTR | 5036 | 16 (5) | 3168 | 62.9 | 21.6 | 1408 | 48.6 |
| Novel exon | 1655 | 81 (28) | 1282 | 77.5 | 19.1 | 411 | 59.8 |
| Novel exon, ambiguous | 543 | 79 (34) | 464 | 85.5 | 30.2 | 268 | 52.3 |
| . | No. found FLA2 . | Average LN coverage FLA2 (median) . | Found in same class FLB1, no. . | Found in same class FLB1, % . | Average PhastCoverage . | PhastCoverage excluding nonoverlapping, no. . | PhastCoverage excluding nonoverlapping, % . |
|---|---|---|---|---|---|---|---|
| Novel gene | 2261 | 270 (34) | 1924 | 85.1 | 33.7 | 968 | 67.0 |
| Small novel gene | 7441 | 79 (28) | 5449 | 73.2 | 6 | 576 | 56.9 |
| Antisense | 553 | 185 (33) | 458 | 82.8 | 30 | 221 | 61.7 |
| Small antisense | 2422 | 84 (28) | 1819 | 75.1 | 12.2 | 403 | 54.7 |
| Novel UTR | 5036 | 16 (5) | 3168 | 62.9 | 21.6 | 1408 | 48.6 |
| Novel exon | 1655 | 81 (28) | 1282 | 77.5 | 19.1 | 411 | 59.8 |
| Novel exon, ambiguous | 543 | 79 (34) | 464 | 85.5 | 30.2 | 268 | 52.3 |
LN indicates length normalized; and UTR, untranslated region.
Given the robust and widespread role for antisense transcription,32 we used our strand-specific RNA-seq data to identify regions of antisense transcriptions within annotated regions. As expected, we found a large number of transcribed novel elements overlapping annotated genes with the majority of these residing within introns (supplemental Table 6). Apart from the number of elements, there does not appear to be any significant difference in the distribution of the large or small antisense elements identified. We could not identify any clear relationship between the expression of the genes and novel antisense elements that overlapped them (data not shown); however, the regulatory relationship between sense/antisense transcripts can be complex and would probably only be definitively revealed by analyzing time-course expression data.33
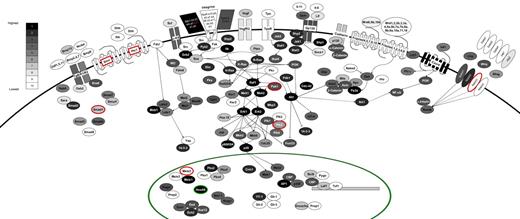
Receptor-ligand expression on leukemia stem cells
We analyzed the expression of selected signaling pathways, which have been implicated in stem cell behavior (Figure 6). The components of each pathway are colored according to their expression within a set of 10 bins (with bin 10 containing all genes not expressed) and with differential expression between FLA2 and FLB1 shown by the red and green colors (FLA2/FLB1 ratio of > 2-fold or < 0.5 in red and green, respectively). Highlighting a possible autocrine activity in L-HSC activity, the following ligand-receptor couples were expressed at the highest levels in our leukemias: Sdf1-Cxcr4; Jagged2-Notch2/1; Osm-Gp130; Scf-cKit; and Bmp15-Tgfb1/2. Vegf-Vegfr2; Wnt6/9b/10b-Frizzled4/5/7/9/10 were expressed but at much lower levels. We also found high expression levels of several integrin genes. Of these, the integrin β2-like gene (Itgb2l) is both highly expressed and differentially expressed between our 2 leukemias (∼ 14-fold higher in FLA2 than FLB1). Besides this exception, no difference in expression pattern could be identified for key members of major signaling pathways thought to participate in hematopoietic stem cell self-renewal. Interestingly and in contrast to normal hematopoietic stem cells, components of the Hedgehog and thrombopoietin pathways (eg, c-mpl) are not expressed. Although the function of the former remains controversial,34-36 c-mpl is critical for HSC quiescence and maintenance.37
Signaling pathways active in FLA2/FLB1 cells. The components of a variety of intracellular signaling pathways are shown, colored according to their expression level. All expressed genes are divided evenly into bins 1 to 9 (1 being most highly expressed) with approximately 1500 genes per bin and all genes that are not expressed (∼ 13k) were placed into bin 10 where expression levels are based on RNA-seq values from FLA2 cells. The color scale corresponding to expression bins is shown in top left of diagram. Genes that are differentially expressed are shown surrounded by a red or green circle (for higher expression in FLA2 or FLB1 cells, respectively) or by red text in the case of Itgb2l (for higher expression in FLA2). Only partial wiring of known signaling pathways is shown to maintain clarity in the diagram.
Signaling pathways active in FLA2/FLB1 cells. The components of a variety of intracellular signaling pathways are shown, colored according to their expression level. All expressed genes are divided evenly into bins 1 to 9 (1 being most highly expressed) with approximately 1500 genes per bin and all genes that are not expressed (∼ 13k) were placed into bin 10 where expression levels are based on RNA-seq values from FLA2 cells. The color scale corresponding to expression bins is shown in top left of diagram. Genes that are differentially expressed are shown surrounded by a red or green circle (for higher expression in FLA2 or FLB1 cells, respectively) or by red text in the case of Itgb2l (for higher expression in FLA2). Only partial wiring of known signaling pathways is shown to maintain clarity in the diagram.
Discussion
This study presents a base pair resolution RNA-seq and microarray analysis of the transcriptome of 2 closely related leukemic cell types generated by the overexpression of Meis1 and HoxA9. We have identified a substantial number of genes that were differentially expressed between FLA2/FLB1 cells, including some that have been implicated in processes, such as cell cycle progression and differentiation. The S100a9/8 genes for instance (up-regulated in FLA2 cell) have been suggested to be used by tumors, via the activation of the mitogen-activated protein kinase p38, to facilitate mobility and invasion.38 Still other genes that are highly expressed specifically in FLA2 have also been found to be highly induced when primitive hematopoietic cells overexpress transcription factors that have been demonstrated to induce the expansion of stem cells (E. Denault, B.T.W., F. Barabé, G.S., unpublished observations, April 2010). This results hint that there may be some commonality between sets of genes that regulate self-renewal of normal and cancerous stem cells. In addition, the GO annotations of the genes that are differentially expressed do show a significant bias and seem to favor differential expression of G-protein-coupled receptors as a general class. It suggests the possibility that perhaps other classes of genes, possibly involved in more fundamental aspects of cell biology, are not as suitable “evolutionary targets” for determining or regulating cell fate. Although determination of cell fate is certainly not as clear-cut as differential activation of a single class of genes, we are unaware of such a bias having been previously reported with respect to differential gene expression in broad GO categories.
Interestingly, a large number of the genes that are highly up-regulated in FLA2 cells (compared with FLB1) belong to the previously characterized cystatin gene family,39,40 consisting of 10 highly related genes located within a cluster on chromosome 16. These genes have not previously been documented to be involved in stem cell behavior or cancer progression and may not even be required for normal cellular growth.39,41 A probable explanation for their up-regulation is that these genes may share a common regulatory element, which is bound by either the overexpressed Meis1/HoxA9 in these cells or by another transcription factor specifically up-regulated in FLA2 cells, as there is no evidence for a viral integration effect near the region of these genes by quantitative RT-PCR (data not shown) or by RNA-seq (supplemental Figure 1).
As shown in Figure 6, the integrin β2-like gene is both highly expressed in our leukemias and shows differential expression (14-fold higher in FLA2). Given the lack of annotation regarding the function of this gene in the context of hematopoiesis, it is not clear whether the differential expression of this gene may have direct relevance to the differences in self-renewal exhibited by the FLA2/FLB1 cells.
Variations in SNV content could also play a role in altering the proteome content. As noted, we located a high confidence, homozygous SNV in the Meis1 gene in the FLB1 cells, which almost certainly derived from the original retroviral transfection. This mutation is within the HR1 required for interaction with PBX1 domain, the loss of which has been demonstrated to cause a longer latency of leukemia development in mice.30 This apparently incidental point mutation provides a potential principle cause for the transcriptional differences between cell types: if the transcriptional complexes of Meis1/HoxA9/Pbx1 differ in stability, efficacy, or localization, this might explain why the transcriptomes of the closely related cells are so different. Moreover, such a scenario would mean that this difference must be “upstream” of whatever changes are induced allowing a higher level of “self renewal” in FLA2 cells, making this system even more interesting as a model for dissecting the mechanisms of L-HSC self-renewal.
As previous studies have noted, transcriptomic characterization by RNA-seq is not only as reproducible as traditional approaches but far more sensitive and generates an extremely rich dataset.42,43 Most remarkably, these data demonstrate that even extremely similar cells can have substantial differences in their transcriptomes not simply at the level of differential expression but also at the level of transcript structures. With respect to the question of how such differences might enable the FLA2 cells to reinitiate leukemias with orders of magnitude greater efficiency than FLB1 cells, we cannot pinpoint a single cause in the many differences that we have found. Despite this, we do now have several interesting possibilities (eg, point mutation in Meis1 in FLB1, differential expression of HSC factors), which we can use to follow-up this initial study to further dissect the molecular mechanism responsible for these self-renewal differences.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank GS laboratory members for valuable comments on this manuscript.
This work was supported by the Canadian Institutes of Health Research (G.S.). B.T.W. was supported by a Cole foundation fellowship.
Authorship
Contribution: B.T.W. and G.S. carried out the experimental design; B.T.W., P.A., A.F., P.C., S.G., N.M., and J.H. carried out the experimental work; B.T.W., M.B., G.B., and J.-R.L. analyzed the data; and B.T.W., K.H., and G.S. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Guy Sauvageau, Institute for Research in Immunology and Cancer, University of Montreal, CP 6128, Downtown Station, Montreal, QC, Canada, H3C 3J7; e-mail:guy.sauvageau@umontreal.ca.