Abstract
Dendritic cells (DCs) play a pivotal role in the pathogenesis of inflammatory disorders, so suppressing the activity of DCs is instrumental in treating such diseases. In the present study, we show that a proteasome inhibitor, bortezomib, suppresses the survival and immunostimulatory function of human plasmacytoid DCs (pDCs) by targeting 2 critical points, intracellular trafficking of nucleic acid-sensingToll-like receptors (TLRs) and endoplasmic reticulum (ER) homeostasis. Among the immune cells in blood, pDCs were the most susceptible to the killing effect of bortezomib. This correlates with a decrease in the spliced form of a transcription factor XBP1, which rescues cells from apoptosis by maintaining ER homeostasis. Bortezomib suppressed the production of interferon-α and interleukin-6 by pDCs activated with a TLR9-stimulating CpG DNA and a TLR7-stimulating influenza virus, which appears to be partially independent of apoptosis. Bortezomib inhibited translocation of TLR9 from the ER to endolysosomes but not of an ER membrane protein, Unc93B1, that delivers TLR9 to endolysosomes. Thus, bortezomib suppresses the activity of pDCs by inhibiting intracellular trafficking of TLRs through disrupting the coordinated translocation of TLRs and Unc93B1 and by disturbing ER homeostasis. This study suggests that proteasome inhibitors may alleviate inflammatory disorders such as lupus and psoriasis that involve pDCs.
Introduction
Dendritic cells (DCs) play a pivotal role in controlling immune responses by linking innate and adaptive arms of the immune system.1 Thus, DCs represent an important target for the treatment of a variety of immune-related disorders.
In humans, DCs are composed of 2 subsets: plasmacytoid DCs (pDCs) and myeloid DCs (mDCs).2 These DCs express different sets of nucleic acid-sensing Toll-like receptors (TLRs): pDCs express TLR7 and TLR9, whereas mDCs express TLR3 and TLR8.3 pDCs are distinguished from other immune cells by their remarkable potency to produce interferon (IFN)-α in response to virus-derived single-stranded RNA or CpG DNA though TLR7 or TLR9, respectively.4 Because of this distinctive capacity, it has been assumed that pDCs play an important role in antiviral immune responses.5 On the other hand, it has been shown that if the nucleic acids are derived from self tissues, pDCs cause inflammatory disorders such as systemic lupus erythematosus (SLE) and psoriasis.6 In SLE, immune complexes containing self DNA or RNA are incorporated into pDCs and induce them to produce IFN-α via TLR97,8 or TLR7,8,9 respectively. It is proposed that such IFN-α production plays a key role in the pathogenesis of SLE.10,11 In psoriasis, aggregated particles composed of self DNA and RNA from damaged epithelial cells and the antimicrobial peptide LL37 are incorporated into pDCs and induce them to produce IFN-α via TLR912 and TLR7,13 respectively, thus contributing to the pathogenesis. Therefore, suppressing IFN-α production by pDCs may represent a novel therapy for these inflammatory disorders in which pDCs are likely to play an important role.
A recent study has suggested that maintenance of endoplasmic reticulum (ER) homeostasis by the transcription factor XBP1 is essential for the development and survival of pDCs, thus representing a possible target for controlling their activity.14 Proper functioning of highly secretory cells such as pDCs depends on the unfolded protein response (UPR), that is, coordinated handling of ER stress caused by a burden of unfolded proteins in the lumen of the ER.15 After sensing unfolded proteins, an ER-resident transmembrane endoribonuclease, IRE1, exhibits unconventional splicing activity on XBP1 mRNA, which results in the conversion of an inactive, unspliced XBP1 (XBP1u) to an active, spliced XBP1 (XBP1s) protein. XBP1s induces the transcription of a broad array of UPR genes that assist in protein synthesis and secretion.16,17 Development of XBP1-deficient pDCs is reduced, likely due to their increased sensitivity to apoptosis induced by ER stress.14
Another important step for the physiologic activity of pDCs is intracellular trafficking of nucleic acid-sensing TLRs from the ER to endolysosomes.18 Recent studies have revealed that a multiple membrane-spanning protein, Unc93B1, physically interacts with nucleic acid-sensing TLRs (TLR3, TLR7, and TLR9) in the ER and delivers them to endolysosomes, where the TLRs transmit an activating signal.19,20 Thus, the interaction between nucleic acid-sensing TLRs and Unc93B1 constitutes another target for controlling the activity of pDCs.
A selective inhibitor of the 26S proteasome, bortezomib, has been established as an effective drug for plasma cell myeloma.21 Although multiple mechanisms have been reported for the antitumor activity of bortezomib,21 growing evidence suggests that the selectivity of bortezomib for myeloma may be explained by increased susceptibility of myeloma cells to ER stress-induced apoptosis,22,23 which is consistent with a crucial role of XBP1 in the development of plasma cells24,25 and in the pathogenesis of myeloma.26 Bortezomib is likely to disturb ER homeostasis of myeloma cells by targeting several points. For example, proteasome inhibitors have been shown to suppress the activity of IRE1 and to stabilize the dominant-negative XBP1u protein, resulting in a decrease in the activity of XBP1s in myeloma cells.22 Proteasome inhibitors also prevent retrograde translocation of misfolded proteins in the ER to the cytosol, resulting in the accumulation of a large amount of misfolded immunoglobulin in the ER of myeloma cells.27 Such overloading of the ER might compromise its physiologic functions. pDCs resemble plasma cells in that both have the developed ER,28 are highly secretory, and depend on ER homeostasis, particularly on XBP1, for their development and survival.14,24,25 Furthermore, coordinated trafficking of the 2 ER-resident proteins, TLR and Unc93B1, is necessary for pDCs to respond to the TLR ligands.20 Therefore, we hypothesized that bortezomib may suppress the activity of pDCs by targeting the 2 critical events in the ER: the UPR and the coordinated function of the ER-resident TLRs and Unc93B1.
In the present study, we investigated the effects of bortezomib on human pDCs. We show that bortezomib inhibits the production of IFN-α by blocking intracellular trafficking of nucleic acid-sensing TLRs at an early stage, and thereafter induces apoptosis of pDCs, which is correlated with the suppression of XBP1 splicing. These results have significant implications for the physiologic mechanisms of survival and activation of pDCs and for the application of proteasome inhibitors to inflammatory disorders in which pDCs play a key role.
Methods
Culture media, reagents, and cell lines
RPMI 1640 (Sigma-Aldrich) supplemented with 10% heat-inactivated fetal calf serum (ThermoTrace), 2mM l-glutamine, penicillin G, streptomycin (Gibco BRL), and 10mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid; Nacalai Tesque) was used for cell culture. Bortezomib, which was provided by Millennium Pharmaceuticals, was dissolved in dimethylsulfoxide at 10mM as a stock solution and was stored at −20°C. Influenza virus (105.3 median tissue culture infective dose/0.2 mL of A/Niigata/05F254/2006), a kind gift from Dr Reiko Saito (Niigata University, Niigata, Japan), was inactivated at 56°C for 30 minutes and added at 0.1% vol/vol to the cell culture. A cell line derived from blastic plasmacytoid dendritic cell neoplasm CAL-1 was described previously by Maeda et al.29 The myeloma cell line RPMI 8226 was obtained from ATCC.
Isolation of pDCs
This study was approved by the Institutional Review Board of the Graduate School of Medicine at Kyoto University and abides by the tenets of the Declaration of Helsinki. Peripheral blood mononuclear cells (PBMCs) were obtained from healthy donors with written informed consent in accordance with the Declaration of Helsinki. pDCs were isolated as described in Kawamura et al.30 In brief, CD4+CD11c−lin− cells were isolated as pDCs using an FACSAria cell sorter (BD Biosciences). Reanalysis of the sorted cells confirmed a purity of more than 98%.
Cell viability assays
PBMCs (2 × 106 cells/2 mL in 12-well culture plates) were cultured with bortezomib for 6 or 24 hours. The cells were stained with the following combinations of monoclonal antibodies (mAbs): fluorescein isothiocyanate (FITC)–conjugated anti-CD3 (BD Biosciences) and PE-conjugated anti-CD56 mAbs (Beckman Coulter); FITC-conjugated CD19 (BD Biosciences) and PE-conjugated anti-BDCA-1 mAbs (Miltenyi Biotec); or FITC-conjugated anti-BDCA-2 (Miltenyi Biotec) and PE-conjugated anti-CD14 mAbs (Beckman Coulter). T cells (CD3+), natural killer (NK) cells (CD3−CD56+), B cells (CD19+), mDCs (BDCA-1+CD19−), pDCs (BDCA-2+), and monocytes (CD14+) were identified by flow cytometry using the FACSCalibur (BD Biosciences). Dead cells were excluded by staining with propidium iodide. Viable cell numbers of each cell population were counted using Flow-Count Fluorospheres (Beckman Coulter) according to the manufacturer's instructions. Purified pDCs (4 × 104 cells/200 μL in round-bottom, 96-well culture plates) were cultured with bortezomib for 3 hours and stimulated with 0.5μM oligodeoxynucleotide 2216 (ODN2216) (CpG-A)31 (Operon Biotechnologies) in the presence of bortezomib for 24 hours. The cells were stained with FITC-conjugated CELL LAB ApoScreen annexin V (Beckman Coulter) and propidium iodide and were analyzed for viability by flow cytometry with the FACSCalibur. Alternatively, pDCs were treated with the pan-caspase inhibitor benzyloxycarbonyl-V-A-D-O-methyl fluoromethyl ketone (50μM Z-VAD-FMK; R&D Systems) together with bortezomib and 0.5μM ODN2216, stained with FITC-conjugated annexin V, and analyzed by flow cytometry. The percentages of specific death induced by bortezomib were calculated as 100 × (experimental death % − spontaneous death %)/(100 − spontaneous death %), in which the percentages of experimental or spontaneous death were defined as the percentages of annexin V–positive cells in the presence or absence of bortezomib, respectively.
RT and real-time PCR
Cells were treated with tunicamycin (Wako Pure Chemical Industries) at 0.4 μg/mL for CAL-1, 0.8 μg/mL for RPMI 8226, or 5 μg/mL for pDCs and T cells to induce ER stress. Total RNA was isolated using the QIAshredder and RNeasy Mini Kit (QIAGEN). First-strand cDNA synthesis was performed with the ReverTra Ace qPCR RT kit (Toyobo). Real-time polymerase chain reaction (PCR) was performed on the Thermal Cycler Dice real-time system (TaKaRa). XBP1 was detected using SYBR Premix Ex Taq (TakaRa) and gene-specific oligonucleotide primers as follows: for the XBP1u gene: 5′-CGAATGAGTGAGCTGGAACA-3′ (forward) and 5′-CTGCAGAGGTGCACGTAGTC-3′ (reverse); for the XBP1s gene: 5′-CGAATGAGTGAGCTGGAACA-3′ (forward) and 5′-CTGCACCTGCTGCGGACT-3′ (reverse).
IFN-α1 and β-glucuronidase (GUS) were detected using TaqMan gene expression assays (Applied Biosystems) and the THUNDERBIRD Probe qPCR mix (Toyobo). Relative quantitation of mRNA expression was performed using the ΔΔCt method. The mRNA expression levels of each gene were normalized to those of GUS.
Analysis of cytokine production by ELISA
Cytokines in the supernatants were analyzed by enzyme-linked immunosorbent assay (ELISA). The following reagents were used: the human IFN-α module set (Bender MedSystems) and the human interleukin-6 (IL-6) ELISA MAX Standard set (BioLegend).
Retroviral transduction
HEK293T cells were cotransfected with retroviral vectors (pMXpuro carrying a green fluorescent protein [GFP])–tagged mouse TLR9 gene32 or pMXneo carrying a GFP-tagged mouse UNC93B1 gene33 ) and retroviral packaging plasmids (pMLVg/p and pVSV-G) using the CalPhos mammalian transfection kit (Clontech). Forty-eight hours after transfection, the supernatant was collected. The mouse B-cell line M1232 was infected with the virus suspension.
Confocal analysis of intracellular trafficking of TLR9 and Unc93B1
M12 cells expressing TLR9-GFP or Unc93B1-GFP were cultured in the absence or presence of 30nM bortezomib for 1 hour, and stimulated with 0.5μM ODN166834 (Operon Biotechnologies) in the absence or presence of 30nM bortezomib for 2 hours. ER-Tracker Red or LysoTracker Red DND-99 (Invitrogen) was added for 30 minutes before the harvest. After fixation with 4% paraformaldehyde for 15 minutes at 37°C, the cells were attached to poly-L-lysine–coated slides and examined with a laser scanning 510 confocal microscope with an αPlan-Fluar 100×/1.45 numeric aperture oil-immersion objective (Carl Zeiss). Data were acquired with the laser scanning microscope software (LSM 510, Version 3.2 SP2 software; Carl Zeiss).
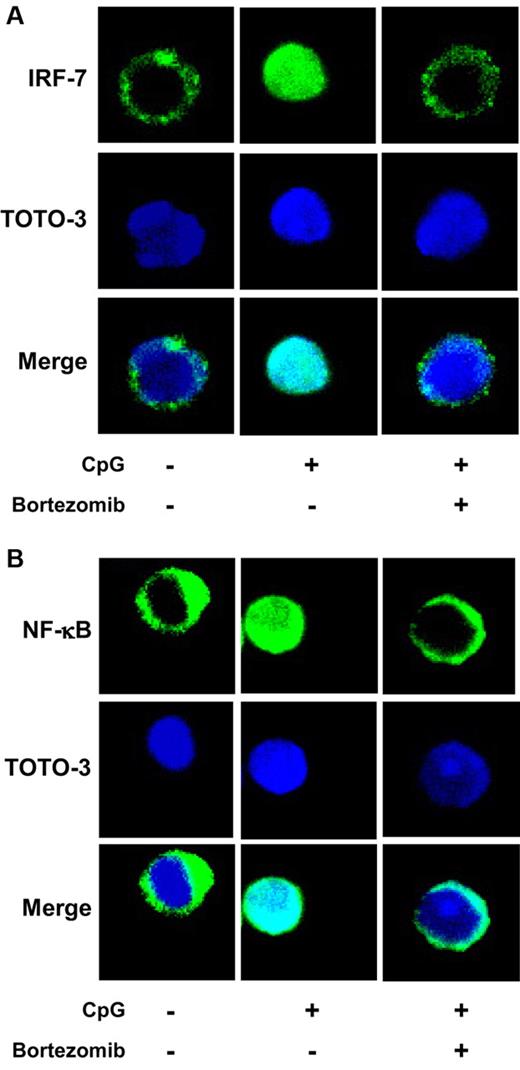
Confocal analysis of nuclear translocation of IRF-7 and NF-κB
Purified pDCs were cultured in the absence or presence of 10nM bortezomib for 3 hours, and were stimulated with 0.5μM ODN2216 in the absence or presence of 10nM bortezomib for 3 hours. After fixation with 2% paraformaldehyde for 15 minutes at 37°C and permeabilization with 100% methanol for 10 minutes at −20°C, the cells were stained with rabbit anti–IFN regulatory factor 7 (anti–IRF-7) or nuclear factor κB (NF-κB) p65 polyclonal antibody (Santa Cruz Biotechnology) and with Alexa Fluor 488-conjugated goat anti–rabbit immunoglobulin G (Invitrogen) as a secondary antibody. Nuclei were identified using TOTO-3 dye (Invitrogen). The cells were attached to slides using a Cytospin centrifuge and examined by confocal microscopy.
Statistical analysis
Data are presented as means ± standard error (SE). Statistical comparisons were performed using paired one-tailed t tests, with a P value < .05 taken to indicate significance.
Results
pDCs are most susceptible to the killing effect of bortezomib among immune cells in blood
We first examined the viability of each mononuclear cell population in the peripheral blood after culture with bortezomib alone. PBMCs were cultured with bortezomib for 6 hours, washed to remove bortezomib, and then cultured for 18 hours. Alternatively, PBMCs were cultured with bortezomib for 24 hours. Thereafter, the viability of each mononuclear cell population was examined by staining the cells with fluorochrome-conjugated mAbs (CD3+ for T cells, CD3−CD56+ for NK cells, CD19+ for B cells, CD14+ for monocytes, BDCA-1+CD19− for mDCs, and BDCA-2+ for pDCs) and propidium iodide. Bortezomib was added at 3-100nM (1.15-38.5 ng/mL), which are the clinically relevant concentrations observed in blood after administration.35,36 The 6-hour exposure to bortezomib mimics the situation in which concentrations are transiently elevated after administration. Resting T cells were the most resistant to the killing effect of bortezomib (Figure 1A), which is consistent with a previous report by Blanco et al.37 NK cells, B cells, and monocytes are more susceptible to the killing effect than are T cells. Bortezomib killed mDCs more than these cells. Notably, pDCs were the most susceptible to the killing effect, and even short-term exposure to 10nM bortezomib greatly reduced their viability after 24 hours.
pDCs are susceptible to the killing effect of bortezomib. (A) PBMCs (2 × 106 cells/2 mL) were cultured without or with the indicated concentrations of bortezomib for 6 hours and washed to remove bortezomib. The cell concentration was then adjusted to 1 × 106/mL and further cultured for 18 hours without bortezomib (Wash [+]). Alternatively, PBMCs (2 × 106 cells/2 mL) were cultured without or with bortezomib for 24 hours (Wash [−]). After staining the PBMCs with mAbs to identify each cell population and with propidium iodide to exclude dead cells, viable cell numbers were counted using Flow-Count Fluorospheres by flow cytometry. Cell numbers in the presence of bortezomib relative to those in the absence of bortezomib were expressed as a percentage. Mo indicates monocytes. (B) Purified pDCs (4 × 104 cells/200 μL) were cultured without or with the indicated concentrations of bortezomib for 3 hours, and stimulated with 0.5μM ODN2216 in the presence of bortezomib for 24 hours. The cells were stained with annexin V and propidium iodide and analyzed by flow cytometry. The numbers indicate the percentages of annexin V and propidium iodide double-negative viable cells. *P < .05; **P < .01; ***P < .001. The data are shown as means ± SE of 3 independent experiments. P values refer to the comparison between the data obtained without bortezomib and those obtained with each concentration of bortezomib.
pDCs are susceptible to the killing effect of bortezomib. (A) PBMCs (2 × 106 cells/2 mL) were cultured without or with the indicated concentrations of bortezomib for 6 hours and washed to remove bortezomib. The cell concentration was then adjusted to 1 × 106/mL and further cultured for 18 hours without bortezomib (Wash [+]). Alternatively, PBMCs (2 × 106 cells/2 mL) were cultured without or with bortezomib for 24 hours (Wash [−]). After staining the PBMCs with mAbs to identify each cell population and with propidium iodide to exclude dead cells, viable cell numbers were counted using Flow-Count Fluorospheres by flow cytometry. Cell numbers in the presence of bortezomib relative to those in the absence of bortezomib were expressed as a percentage. Mo indicates monocytes. (B) Purified pDCs (4 × 104 cells/200 μL) were cultured without or with the indicated concentrations of bortezomib for 3 hours, and stimulated with 0.5μM ODN2216 in the presence of bortezomib for 24 hours. The cells were stained with annexin V and propidium iodide and analyzed by flow cytometry. The numbers indicate the percentages of annexin V and propidium iodide double-negative viable cells. *P < .05; **P < .01; ***P < .001. The data are shown as means ± SE of 3 independent experiments. P values refer to the comparison between the data obtained without bortezomib and those obtained with each concentration of bortezomib.
We further examined whether bortezomib also reduces the viability of pDCs stimulated with the TLR9 ligand ODN2216 (CpG-A). Because pDCs cannot be unequivocally identified among PBMCs after stimulation with CpG ODN due to the change of their surface phenotype,38 we purified pDCs, stimulated them with ODN2216 in the absence or presence of different concentrations of bortezomib for 24 hours, and analyzed their viability with annexin V and propidium iodide staining (Figure 1B). Whereas the majority of pDCs underwent apoptosis without stimulation, as shown in previous studies,28,39 stimulation with ODN2216 greatly improved the viability. A bortezomib concentration of 30nM or above strongly induced apoptosis of ODN2216-stimulated pDCs.
These data indicate that, among the different populations of PBMCs, bortezomib has the strongest killing effect on pDCs in a resting condition and also induces apoptosis of pDCs stimulated with CpG ODN.
Inhibition of XBP1 splicing correlates with the induction of apoptosis of pDCs by bortezomib
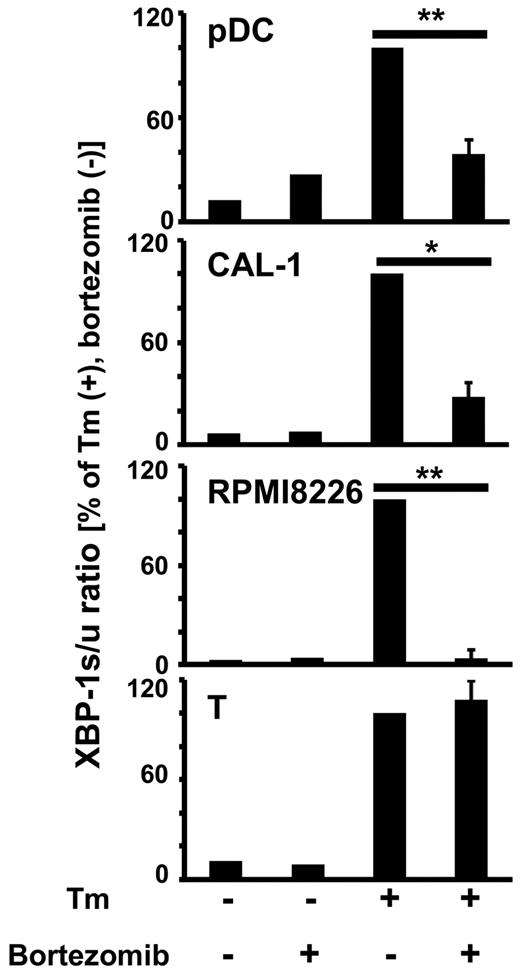
Because it has been shown that XBP1 is essential for the development of plasma cells24,25 and pDCs,14 and that bortezomib disrupts the UPR by inhibiting the generation of spliced XBP1 (XBP1s), the active form of XBP1, in plasma cells,22 we examined whether bortezomib also inhibits the generation of XBP1s mRNA in pDCs. We cultured primary pDCs, a cell line derived from blastic plasmacytoid dendritic cell neoplasm CAL-1,29 a myeloma cell line RPMI 8226, and T cells in the absence or presence of bortezomib and tunicamycin, an inhibitor of N-linked glycosylation known to induce ER stress and the UPR. The amounts of XBP1s mRNA and XBP1u mRNA were quantitated by real-time reverse transcription (RT)–PCR, and the ratios of XBP1s to XBP1u were calculated (Figure 2). The addition of tunicamycin increased the amounts of XBP1s mRNA relative to those of XBP1u mRNA in all of the cell types. The addition of bortezomib suppressed the generation of XBP1s mRNA induced by tunicamycin in pDCs, CAL-1, and RPMI 8226, whereas bortezomib did not decrease XBP1s mRNA in T cells, which are resistant to the killing effect of bortezomib (Figure 1A). Thus, apoptosis of pDCs is correlated with the decrease in XBP1s, suggesting that disruption of ER homeostasis by bortezomib results in apoptosis of pDCs as well as of plasma cells.22
Bortezomib inhibits XBP1 splicing in pDCs. Purified pDCs, the pDC tumor cell line CAL-1, the myeloma cell line RPMI 8226, or resting T cells were cultured without or with 100nM bortezomib for 1 hour, and 5 μg/mL tunicamycin (Tm) was added 4 hours before harvest. The expression levels of XBP1u and XBP1s mRNA were measured by real-time RT-PCR and normalized to those of GUS. The XBP1s/XBP1u ratios were calculated. The data are normalized to the value obtained with tunicamycin in the absence of bortezomib. *P < .05; **P < .01. The data are shown as means ± SE of 3 (pDC) or 4 (CAL-1, RPMI8226, T) independent experiments.
Bortezomib inhibits XBP1 splicing in pDCs. Purified pDCs, the pDC tumor cell line CAL-1, the myeloma cell line RPMI 8226, or resting T cells were cultured without or with 100nM bortezomib for 1 hour, and 5 μg/mL tunicamycin (Tm) was added 4 hours before harvest. The expression levels of XBP1u and XBP1s mRNA were measured by real-time RT-PCR and normalized to those of GUS. The XBP1s/XBP1u ratios were calculated. The data are normalized to the value obtained with tunicamycin in the absence of bortezomib. *P < .05; **P < .01. The data are shown as means ± SE of 3 (pDC) or 4 (CAL-1, RPMI8226, T) independent experiments.
Bortezomib suppresses cytokine production by pDCs stimulated with CpG ODN or influenza virus
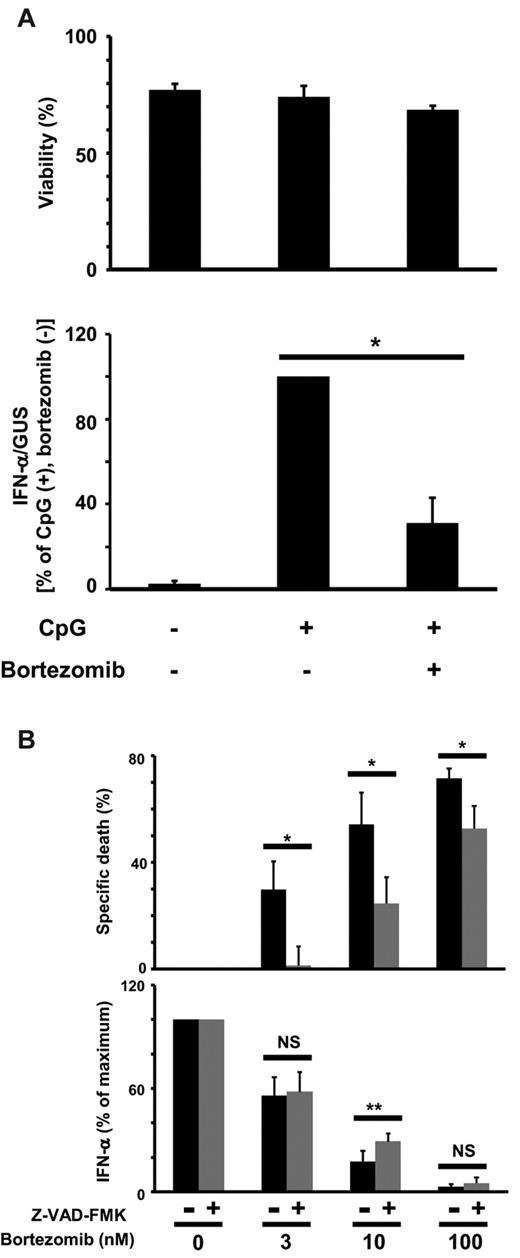
Next we examined whether bortezomib suppresses cytokine production by pDCs stimulated with ODN2216 (a TLR9 ligand) or influenza virus (a TLR7 ligand). We pretreated PBMCs with different concentrations of bortezomib for 6 hours, washed the cells to remove bortezomib, and cultured them with the TLR ligands for 24 hours. Alternatively, we pretreated PBMCs with bortezomib for 6 hours, added the TLR ligands without removing bortezomib, and cultured the cells for 24 hours. The concentrations of IFN-α in the supernatants were then measured by ELISA. Because the absolute concentrations were variable depending on the donors, the levels of cytokine concentrations were normalized to the maximum value obtained in the absence of bortezomib. Pretreatment with bortezomib suppressed the production of IFN-α induced by ODN2216 or influenza virus in a dose-dependent manner (Figure 3A, Wash [+]). The addition of bortezomib during the whole period of culture further suppressed the production of IFN-α (Figure 3A, Wash [−]). These data indicate that bortezomib strongly suppresses the production of IFN-α by pDCs stimulated with the TLR9 or TLR7 ligand in a dose- and time-dependent manner.
Bortezomib suppresses cytokine production by pDCs stimulated with ODN2216 or influenza virus. (A) PBMCs (1 × 106/mL) were cultured without or with the indicated concentrations of bortezomib for 6 hours and washed to remove bortezomib. The cell concentration was then adjusted to 1 × 106/mL and further cultured for 24 hours in the presence of 0.5μM ODN2216 or influenza virus without bortezomib (Wash [+]). Alternatively, PBMCs (1 × 106/mL) were cultured without or with bortezomib for 6 hours, and cultured for 24 hours in the presence of ODN2216 or influenza virus without removing bortezomib (Wash [−]). Concentrations of IFN-α in the supernatants were measured by ELISA. The data are normalized to the value obtained without bortezomib. *P < .05; **P < .01; ***P < .001. The data are shown as means ± SE of 3 independent experiments. P values refer to the comparison between the data obtained without bortezomib and those obtained with each concentration of bortezomib. The means and ranges of absolute concentrations are as follows: ODN2216 (Wash [−]) 4292 pg/mL (1674-9455 pg/mL); influenza (Wash [−]) 1278 pg/mL (1008-1538 pg/mL). (B) Purified pDCs (2 × 105/mL) were cultured without or with the indicated concentrations of bortezomib for 3 hours, and cultured for 24 hours in the presence of 0.5μM ODN2216 without removing bortezomib. Concentrations of IFN-α and IL-6 in the supernatants were measured by ELISA. **P < .01; ***P < .001. The data are shown as means ± SE of 3 independent experiments. The means and ranges of absolute concentrations were as follows: IFN-α, 74 312 pg/mL (48 118-92 759 pg/mL); IL-6, 3904 pg/mL (1970-6343 pg/mL).
Bortezomib suppresses cytokine production by pDCs stimulated with ODN2216 or influenza virus. (A) PBMCs (1 × 106/mL) were cultured without or with the indicated concentrations of bortezomib for 6 hours and washed to remove bortezomib. The cell concentration was then adjusted to 1 × 106/mL and further cultured for 24 hours in the presence of 0.5μM ODN2216 or influenza virus without bortezomib (Wash [+]). Alternatively, PBMCs (1 × 106/mL) were cultured without or with bortezomib for 6 hours, and cultured for 24 hours in the presence of ODN2216 or influenza virus without removing bortezomib (Wash [−]). Concentrations of IFN-α in the supernatants were measured by ELISA. The data are normalized to the value obtained without bortezomib. *P < .05; **P < .01; ***P < .001. The data are shown as means ± SE of 3 independent experiments. P values refer to the comparison between the data obtained without bortezomib and those obtained with each concentration of bortezomib. The means and ranges of absolute concentrations are as follows: ODN2216 (Wash [−]) 4292 pg/mL (1674-9455 pg/mL); influenza (Wash [−]) 1278 pg/mL (1008-1538 pg/mL). (B) Purified pDCs (2 × 105/mL) were cultured without or with the indicated concentrations of bortezomib for 3 hours, and cultured for 24 hours in the presence of 0.5μM ODN2216 without removing bortezomib. Concentrations of IFN-α and IL-6 in the supernatants were measured by ELISA. **P < .01; ***P < .001. The data are shown as means ± SE of 3 independent experiments. The means and ranges of absolute concentrations were as follows: IFN-α, 74 312 pg/mL (48 118-92 759 pg/mL); IL-6, 3904 pg/mL (1970-6343 pg/mL).
We also examined the effect of bortezomib on the production of IFN-α and IL-6 by purified pDCs stimulated with ODN2216. We pretreated purified pDCs with different concentrations of bortezomib for 3 hours, added ODN2216 without removing bortezomib, and cultured the cells for 24 hours. The concentrations of IFN-α and IL-6 in the supernatants were then measured by ELISA. Bortezomib suppressed the production of both IFN-α and IL-6 by purified pDCs in a dose-dependent manner (Figure 3B).
To examine whether the suppression of cytokine production is due to the induction of apoptosis, we stimulated pDCs with ODN2216 in the absence or presence of bortezomib for 4 hours. Viability was then measured with annexin V/propidium iodide staining, and the amounts of IFN-α mRNA were measured by real-time RT-PCR. Whereas viability did not decrease during the culture, bortezomib strongly suppressed the transcription of IFN-α mRNA (Figure 4A). We also cultured pDCs with ODN2216 and different concentrations of bortezomib in the absence or presence of the pan-caspase inhibitor Z-VAD-FMK. The percentages of apoptosis specifically induced by bortezomib were then calculated and the concentrations of IFN-α in the supernatants measured by ELISA. Although Z-VAD-FMK significantly suppressed apoptosis of pDCs induced by each concentration (3, 10, 100nM) of bortezomib, the recovery of IFN-α production by Z-VAD-FMK was marginal and was significant only at 10nM bortezomib (Figure 4B). Based on these data, we concluded that bortezomib suppresses IFN-α production by pDCs in a partly apoptosis-independent manner.
Relationship between viability and IFN-α production by pDCs. (A) Purified pDCs were cultured with 10nM bortezomib for 2 hours and 0.5μM ODN2216 was added. The cells were harvested after 4 hours, stained with FITC-conjugated annexin V, and analyzed for viability by flow cytometry (top panel). IFN-α mRNA was quantitated by real-time RT-PCR, and the expression levels were normalized to those of GUS (bottom panel). The data are normalized to the value obtained with ODN2216 in the absence of bortezomib. *P < .05. The data are shown as means ± SE of 3 independent experiments. (B) After purified pDCs (1 × 105/mL) were cultured without or with 50μM Z-VAD-FMK for 1 hour, the indicated concentrations of bortezomib were added. After 6 hours, 0.5μM ODN2216 was added and the cells and supernatants were harvested 18 hours later. The cells were stained with FITC-conjugated annexin V and analyzed by flow cytometry. The percentages of cell death specifically induced by bortezomib were calculated (top panel). Concentrations of IFN-α in the supernatants were measured by ELISA (bottom panel). The data are normalized to the value obtained without bortezomib. *P < .05; **P < .01; NS, not significant. The data are shown as means ± SE of 6 independent experiments.
Relationship between viability and IFN-α production by pDCs. (A) Purified pDCs were cultured with 10nM bortezomib for 2 hours and 0.5μM ODN2216 was added. The cells were harvested after 4 hours, stained with FITC-conjugated annexin V, and analyzed for viability by flow cytometry (top panel). IFN-α mRNA was quantitated by real-time RT-PCR, and the expression levels were normalized to those of GUS (bottom panel). The data are normalized to the value obtained with ODN2216 in the absence of bortezomib. *P < .05. The data are shown as means ± SE of 3 independent experiments. (B) After purified pDCs (1 × 105/mL) were cultured without or with 50μM Z-VAD-FMK for 1 hour, the indicated concentrations of bortezomib were added. After 6 hours, 0.5μM ODN2216 was added and the cells and supernatants were harvested 18 hours later. The cells were stained with FITC-conjugated annexin V and analyzed by flow cytometry. The percentages of cell death specifically induced by bortezomib were calculated (top panel). Concentrations of IFN-α in the supernatants were measured by ELISA (bottom panel). The data are normalized to the value obtained without bortezomib. *P < .05; **P < .01; NS, not significant. The data are shown as means ± SE of 6 independent experiments.
Bortezomib inhibits trafficking of TLR9 but not of Unc93B1 from the ER to endolysosomes induced by CpG ODN
We investigated the mechanisms by which bortezomib suppresses cytokine production by pDCs. The earliest event leading to CpG ODN-induced IFN-α production was endocytosis of CpG ODN by pDCs. Thus, we examined whether bortezomib inhibits uptake of CpG ODN by pDCs. We pretreated pDCs with bortezomib for 3 hours, and then added FITC-conjugated ODN2216. After 90 minutes, we examined the intracellular localization of ODN2216 by confocal microscopy (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Whereas pDCs kept on ice did not endocytose ODN2216, pDCs cultured at 37°C did. Bortezomib did not inhibit the endocytosis of ODN2216. Thus, bortezomib targets a signaling pathway(s) farther downstream.
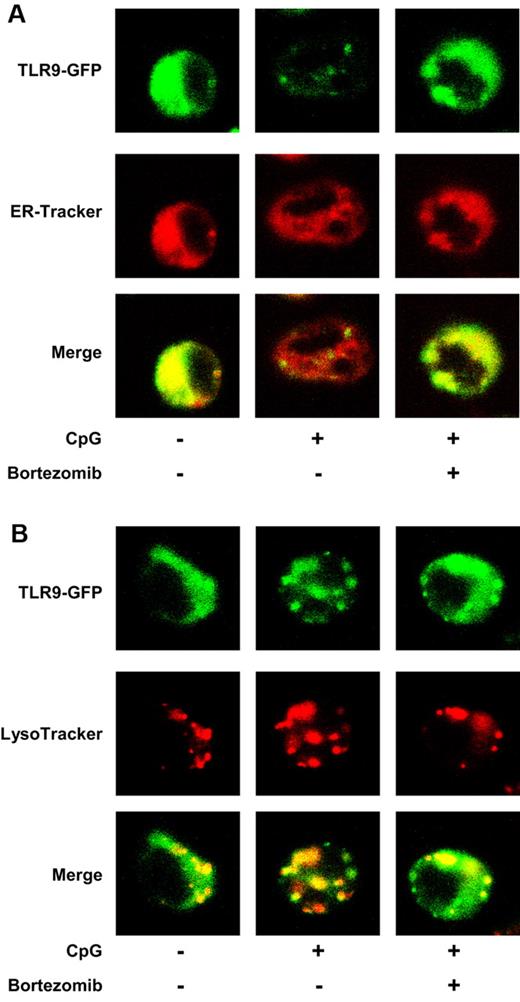
Upon stimulation with CpG ODN, TLR9 rapidly moves from the ER to endolysosomes.18 Thus, we next examined whether bortezomib inhibits the intracellular trafficking of TLR9. We stimulated the mouse B-cell line M12 expressing TLR9-GFP with ODN1668 in the absence or presence of bortezomib, and examined whether TLR9 colocalizes with an ER marker (ER-Tracker) or a lysosomal marker (LysoTracker) by confocal microscopy. Whereas TLR9 was located in the ER (Figure 5A) but not in the endolysosomes (Figure 5B) without stimulation, TLR9 left the ER (Figure 5A) and moved to the endolysosomes (Figure 5B) after stimulation with CpG ODN. In the presence of bortezomib, TLR9 remained in the ER (Figure 5A) and did not move to endolysosomes (Figure 5B) after stimulation. We also used a human pDC line, CAL-1, and obtained similar results: bortezomib inhibited the trafficking of TLR9 from the ER to endolysosomes induced by ODN2216 (supplemental Figure 2).
Bortezomib inhibits the trafficking of TLR9 from the ER to endolysosomes induced by CpG ODN. The mouse B-cell line M12 expressing TLR9-GFP was cultured in the absence or presence of 30nM bortezomib for 1 hour and cultured with ODN1668 in the absence or presence of bortezomib for 2 hours. ER-Tracker (A) and LysoTracker (B) were added during the last 30 minutes. The cells were observed by confocal microscopy. The data are representative of 4 experiments.
Bortezomib inhibits the trafficking of TLR9 from the ER to endolysosomes induced by CpG ODN. The mouse B-cell line M12 expressing TLR9-GFP was cultured in the absence or presence of 30nM bortezomib for 1 hour and cultured with ODN1668 in the absence or presence of bortezomib for 2 hours. ER-Tracker (A) and LysoTracker (B) were added during the last 30 minutes. The cells were observed by confocal microscopy. The data are representative of 4 experiments.
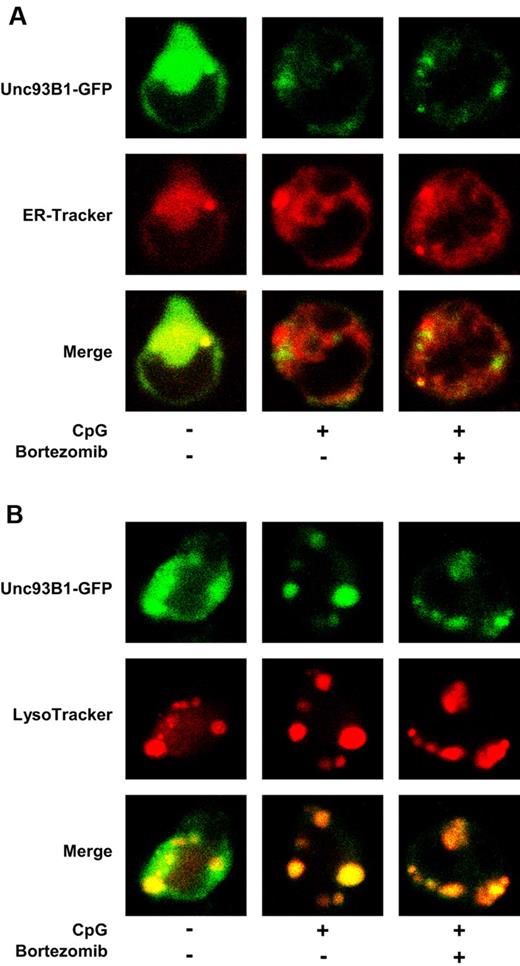
Recent studies have revealed that a multiple membrane-spanning protein, Unc93B1, physically interacts with nucleic acid-sensing TLRs in the ER and delivers them to endolysosomes upon stimulation with a TLR ligand.19,20 Thus, we examined whether bortezomib inhibits the trafficking of TLR9 by inhibiting that of Unc93B1. We stimulated M12 expressing Unc93B1-GFP with ODN1668 in the absence or presence of bortezomib, and examined whether Unc93B1 colocalizes with an ER marker (ER-Tracker) or a lysosomal marker (LysoTracker) by confocal microscopy. Whereas Unc93B1 was located in the ER (Figure 6A) but not in endolysosomes (Figure 6B) without stimulation, Unc93B1 left the ER (Figure 6A) and moved to endolysosomes (Figure 6B) after stimulation with CpG1668. Notably, the translocation of Unc93B1 from the ER to endolysosomes was not abrogated by bortezomib (Figure 6A-B).
Bortezomib does not inhibit the trafficking of Unc93B1 from the ER to endolysosomes induced by CpG ODN. M12 expressing Unc93B1-GFP was cultured in the absence or presence of 30nM bortezomib for 1 hour and cultured with 0.5μM ODN1668 in the absence or presence of bortezomib for 2 hours. ER-Tracker (A) and LysoTracker (B) were added during the last 30 minutes. The cells were observed by confocal microscopy. The data are representative of 4 experiments.
Bortezomib does not inhibit the trafficking of Unc93B1 from the ER to endolysosomes induced by CpG ODN. M12 expressing Unc93B1-GFP was cultured in the absence or presence of 30nM bortezomib for 1 hour and cultured with 0.5μM ODN1668 in the absence or presence of bortezomib for 2 hours. ER-Tracker (A) and LysoTracker (B) were added during the last 30 minutes. The cells were observed by confocal microscopy. The data are representative of 4 experiments.
These data indicate that bortezomib inhibits the trafficking of TLR9 but not of Unc93B1 from the ER to endolysosomes upon stimulation with CpG ODN. This suggests that bortezomib suppresses the immunostimulatory functions of TLR-triggered pDCs by disrupting the coordinated trafficking of TLR and Unc93B1. In addition, because the inhibition of TLR9 trafficking was observed during the early period of culture (3 hours after adding bortezomib), when apoptosis had not ensued, bortezomib is likely to suppress the immunostimulatory function of pDCs independently of the induction of apoptosis (at least in part).
Bortezomib inhibits nuclear translocation of IRF-7 and NF-κB in pDCs stimulated with CpG ODN
Stimulation of pDCs with CpG ODN induces the nuclear translocation of 2 major transcription factors, IRF-7 and NF-κB, which induce the production of IFN-α and proinflammatory cytokines (TNF-α and IL-6), respectively, at the final step of TLR signaling.40 Thus, we examined whether bortezomib inhibits the nuclear translocation of IRF-7 and NF-κB induced by ODN2216 in pDCs consistently with the inhibition of intracellular trafficking of TLR9 (Figure 5) and the suppression of IFN-α and IL-6 production (Figure 3). We pretreated pDCs with bortezomib for 3 hours, stimulated them with ODN2216 for 3 hours, and then examined the localization of IRF-7 (Figure 7A) and NF-κB (Figure 7B) by confocal microscopy. Whereas IRF-7 and NF-κB are located in the cytoplasm in untreated pDCs, both of the transcription factors moved to the nucleus after stimulation with ODN2216. Bortezomib inhibited the nuclear translocation of IRF-7 and NF-κB. These data indicate that the inhibition of TLR9 trafficking by bortezomib leads to the abrogation of the nuclear translocation of IRF-7 and NF-κB.
Bortezomib inhibits the nuclear translocation of IRF-7 and NF-κB in pDCs. Purified pDCs were cultured in the absence or presence of 10nM bortezomib for 3 hours and stimulated with 0.5μM ODN2216 for 3 hours. The cells were stained with rabbit anti–IRF-7 (A) or NF-κB p65 (B) and with Alexa Fluor 488–conjugated goat anti–rabbit immunoglobulin G as a secondary antibody. Nuclei were identified using TOTO-3 dye. The data are representative of 3 experiments.
Bortezomib inhibits the nuclear translocation of IRF-7 and NF-κB in pDCs. Purified pDCs were cultured in the absence or presence of 10nM bortezomib for 3 hours and stimulated with 0.5μM ODN2216 for 3 hours. The cells were stained with rabbit anti–IRF-7 (A) or NF-κB p65 (B) and with Alexa Fluor 488–conjugated goat anti–rabbit immunoglobulin G as a secondary antibody. Nuclei were identified using TOTO-3 dye. The data are representative of 3 experiments.
Discussion
Because of the proposed involvement of pDCs in the pathogenesis of several inflammatory disorders, it is of great importance to find reagents that modulate pDC functions. pDCs and plasma cells share a distinctive property, in that both types of cells have a highly developed ER and produce vast amounts of secretory proteins: IFN-α and immunoglobulin, respectively. Therefore, we hypothesized that the proteasome inhibitor bortezomib, which kills myeloma cells partly due to its effect on ER homeostasis, may affect the viability and function of pDCs. The present study had 2 novel findings. First, bortezomib suppresses the immunostimulatory activity of pDCs by disrupting the coordinated translocation of TLR9 and an ER-resident protein, Unc93B1. Second, bortezomib induces apoptosis of pDCs apparently by disturbing ER homeostasis maintained through the activation of XBP1. This is the first study showing the pharmacologic disruption of the coordinated translocation of nucleic acid–sensing TLRs and Unc93B1, and it sheds new light on the molecular mechanisms by which pDCs perform their immune functions and on the mechanisms by which bortezomib executes its immunosuppressive activity.
We first examined the effect of bortezomib on the viability of pDCs using total PBMCs (Figure 1A) or purified pDCs (Figure 1B). Using total PBMCs, we found that of the different immune cell types in blood, pDCs were the most prone to die even after a 6-hour pretreatment with 10nM bortezomib and subsequent culture without bortezomib for 18 hours. Because the majority of purified pDCs spontaneously underwent apoptosis after 24 hours, as shown in previous studies,28,39 we examined the effect of bortezomib on the viability of purified pDCs (2 × 105/mL) under stimulation with CpG ODN, and found that most of them died after exposure to bortezomib concentrations of 30nM or more. There have been conflicting reports concerning the effect of bortezomib on the viability of pDCs. Kukreja et al reported that the majority of purified pDCs undergo apoptosis after culture either with 100nM bortezomib for 48 hours or with 10nM bortezomib for 24 hours.41 In contrast, Chauhan et al reported that the majority of 1 × 106 purified pDCs are viable even after culture with 20nM bortezomib for 24 hours.42 Although the reason for such differences in the viability of pDCs in different studies is not known, it appears that Chauhan et al cultured pDCs at a much higher cell concentration than we did in this study. Such a difference in culture conditions might have resulted in the observed differences in the effect of bortezomib. In any event, our data using total PBMCs and purified pDCs clearly show that pDCs are prone to die after exposure to bortezomib.
Highly secretory cells such as plasma cells24,25 and exocrine gland acinar cells43 depend on the UPR for their development and survival, as evidenced in XBP1-deficient conditions. It has been shown that proteasome inhibitors target XBP1 through suppressing the activity of IRE1.22 pDCs are also highly secretory and depend on XBP1 for their development and survival.14 Thus, we examined whether bortezomib suppresses the generation of XBP1s in pDCs. We found that bortezomib strongly suppressed XBP1 splicing in bortezomib-susceptible pDCs and in myeloma cells, but not in bortezomib-resistant resting T cells. These data, together with a previous report by Iwakoshi et al,14 suggest that the suppression of XBP1 splicing by bortezomib leads to the apoptosis of pDCs.
We next examined the effect of bortezomib on the cardinal feature of pDCs: the production of IFN-α in response to TLR7 and TLR9 ligands. We also examined the production of the proinflammatory cytokine IL-6, because IFN-α and IL-6 are induced by different transcription factors (IRF-744 and NF-κB,40 respectively). Bortezomib suppressed the production of IFN-α and IL-6 by pDCs stimulated with the TLR9 ligand ODN2216 (CpG-A), and also the production of IFN-α induced by a TLR7 ligand influenza virus, in a dose-dependent manner. Bortezomib appears to suppress the cytokine production by pDCs independently of the induction of apoptosis, at least in part, because: (1) bortezomib suppressed the trafficking of TLR9 and the induction of IFN-α mRNA at an early time point, when apoptosis of pDCs had not ensued; and (2) a pan-caspase inhibitor, Z-VAD-FMK, rescued the IFN-α production only marginally, whereas the reagent significantly rescued pDCs from apoptosis. However, we cannot exclude the possibility that the diminution of cytokine production reflects early apoptotic signaling. For example, it has been shown that bortezomib induces the generation of reactive oxygen species45 and the activation of c-Jun NH2-terminal kinase46 after ER stress, leading to the mitochondrial apoptotic pathway. Such early apoptotic signals might affect functions of pDCs.
Next we examined the mechanisms by which bortezomib suppresses cytokine production by pDCs. Observation by confocal microscopy showed that bortezomib inhibits the intracellular trafficking of TLR9 from the ER to endolysosomes. Notably, however, bortezomib did not inhibit the trafficking of Unc93B1, which has been reported to deliver nucleic acid-sensing TLRs from the ER to endolysosomes.20 These data suggest that bortezomib inhibits the responses of pDCs to nucleic acids by disrupting the coordinated movement of TLRs and Unc93B1. We confirmed that such inhibition of the trafficking of TLR9 resulted in the abrogation of nuclear translocation of IRF-7 and NF-κB, which corresponds to the suppression of IFN-α44 and IL-640 production, respectively.
Because the precise molecular mechanisms by which Unc93B1 delivers TLRs from the ER to endolysosomes remain to be determined, the mechanism by which bortezomib disrupts the coordinated trafficking of TLR9 and Unc93B1 is not clear. It is tempting to speculate that the accumulation of misfolded proteins in the ER caused by bortezomib may disturb proper functions of ER-resident proteins responsible for the trafficking of TLR9 and Unc93B1. We have recently shown that wild-type and D34A mutant Unc93B1 preferentially associate and translocate with TLR9 and TLR7, respectively,33 implying that an ER protein not yet identified interacts with the N-terminal region of Unc93B1 and facilitates the trafficking of Unc93B1-TLR complexes to endolysosomes. Such a mechanism might be disarranged by the overwhelming ER stress caused by bortezomib.
It has been proposed that IFN-α from pDCs stimulated with endogenous nucleic acids plays a key role in the pathogenesis of several autoimmune or inflammatory disorders.47 First, immune complexes composed of DNA/anti-DNA autoantibodies7,8 or ribonucleoprotein/anti-ribonucleoprotein autoantibodies8,9 are incorporated into pDCs and stimulate them to produce IFN-α by triggering TLR9 and TLR7, respectively. Such IFN-α is implicated in the pathogenesis of SLE. Intriguingly, a study using Unc93B1 3d mutant mice, in which Unc93B1 is incapable of binding to TLRs,20 has shown that nucleic acid–sensing TLRs are required for the optimal production of autoantibodies in lupus-prone strains.48 Thus, the immune complexes and pDCs appear to constitute a positive feedback loop mediated by the TLR-Unc93B1 interaction. Proteasome inhibitors may disrupt such a vicious cycle by targeting pDCs, resulting in the alleviation of SLE. Second, aggregates composed of the antimicrobial peptide LL37 and self DNA12 or RNA13 released from damaged cells are incorporated into pDCs and stimulate them to produce IFN-α by triggering TLR9 and TLR7, respectively, and this IFN-α is implicated in the pathogenesis of psoriasis. Bortezomib may alleviate psoriasis by targeting pDCs. Furthermore, pDCs may be involved in the pathogenesis of type 1 diabetes49 and chronic graft-versus-host disease.50 These findings suggest that proteasome inhibitors may be exploited for the treatment of these autoimmune or inflammatory disorders through their suppressive activity on pDCs. The second generation of proteasome inhibitors that have less neurotoxicity than bortezomib and are thus more suitable for long-term use may be applicable to such chronic inflammatory disorders.
In conclusion, the results of this study suggest that bortezomib suppresses the activity of pDCs by disrupting the coordinated trafficking of nucleic acid-sensing TLRs and Unc93B1 from the ER to endolysosomes and by suppressing the UPR. Bortezomib or a next generation of proteasome inhibitors may therefore be instrumental in treating pDC-mediated, IFN-α-driven inflammatory disorders.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Millennium Pharmaceuticals for providing bortezomib, Dr Reiko Saito for providing influenza virus, and Keiko Fukunaga for her excellent technical assistance.
This study was supported by research funding from Ministry of Education, Culture, Sports, Science, and Technology of Japan (17016034).
Authorship
Contribution: M.H. performed research and cowrote the manuscript; N.K. designed research, analyzed and interpreted data, and cowrote the manuscript; T.K. contributed vital experimental designs and techniques; H.F. performed research; A.T.-K. contributed vital experimental designs and techniques; R.F. and K.M. contributed vital analytical tools; T.M. and S.K. contributed vital new reagents; and Y.M. and T.U. supervised the whole project.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Norimitsu Kadowaki, Department of Hematology and Oncology, Graduate School of Medicine, Kyoto University, 54 Shogoin Kawara-cho, Sakyo-ku, Kyoto 606-8507, Japan; e-mail: kadowaki@kuhp.kyoto-u.ac.jp.

![Figure 1. pDCs are susceptible to the killing effect of bortezomib. (A) PBMCs (2 × 106 cells/2 mL) were cultured without or with the indicated concentrations of bortezomib for 6 hours and washed to remove bortezomib. The cell concentration was then adjusted to 1 × 106/mL and further cultured for 18 hours without bortezomib (Wash [+]). Alternatively, PBMCs (2 × 106 cells/2 mL) were cultured without or with bortezomib for 24 hours (Wash [−]). After staining the PBMCs with mAbs to identify each cell population and with propidium iodide to exclude dead cells, viable cell numbers were counted using Flow-Count Fluorospheres by flow cytometry. Cell numbers in the presence of bortezomib relative to those in the absence of bortezomib were expressed as a percentage. Mo indicates monocytes. (B) Purified pDCs (4 × 104 cells/200 μL) were cultured without or with the indicated concentrations of bortezomib for 3 hours, and stimulated with 0.5μM ODN2216 in the presence of bortezomib for 24 hours. The cells were stained with annexin V and propidium iodide and analyzed by flow cytometry. The numbers indicate the percentages of annexin V and propidium iodide double-negative viable cells. *P < .05; **P < .01; ***P < .001. The data are shown as means ± SE of 3 independent experiments. P values refer to the comparison between the data obtained without bortezomib and those obtained with each concentration of bortezomib.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/2/10.1182_blood-2010-05-284737/4/m_zh89991063850001.jpeg?Expires=1765894021&Signature=Rga0D8Lr2q4TAbzVXd-7Xy9M9OpEdkav86xgoheMFmtvlKOBiiHP9wAJHXgJiGCq8JiNiDsYUFETTjE2YJ7A5GHci0B4RD-wIcjwkc35Hx7fjHY377Uk1xG1csm~O5cyQsqF6oenqyQjbSIGzRWSi~9UGS2NjfSD1CSp1bsfwdMTAlj2hHPSPh5WdrFpvAeZLXZWgXWd2NNBnao1yIKpNwJhEE148~yjuEVc8LNDJeKb7Rrcs9KDhWqCsRMuwf65d08I12tbqTbF~3cEdM0yDZ6VGUa86drcYAS2zCrpe-lEXvZmJWuKqEw8odUY-5lH978yBLauvnWZ3OZUUSl7DQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. Bortezomib suppresses cytokine production by pDCs stimulated with ODN2216 or influenza virus. (A) PBMCs (1 × 106/mL) were cultured without or with the indicated concentrations of bortezomib for 6 hours and washed to remove bortezomib. The cell concentration was then adjusted to 1 × 106/mL and further cultured for 24 hours in the presence of 0.5μM ODN2216 or influenza virus without bortezomib (Wash [+]). Alternatively, PBMCs (1 × 106/mL) were cultured without or with bortezomib for 6 hours, and cultured for 24 hours in the presence of ODN2216 or influenza virus without removing bortezomib (Wash [−]). Concentrations of IFN-α in the supernatants were measured by ELISA. The data are normalized to the value obtained without bortezomib. *P < .05; **P < .01; ***P < .001. The data are shown as means ± SE of 3 independent experiments. P values refer to the comparison between the data obtained without bortezomib and those obtained with each concentration of bortezomib. The means and ranges of absolute concentrations are as follows: ODN2216 (Wash [−]) 4292 pg/mL (1674-9455 pg/mL); influenza (Wash [−]) 1278 pg/mL (1008-1538 pg/mL). (B) Purified pDCs (2 × 105/mL) were cultured without or with the indicated concentrations of bortezomib for 3 hours, and cultured for 24 hours in the presence of 0.5μM ODN2216 without removing bortezomib. Concentrations of IFN-α and IL-6 in the supernatants were measured by ELISA. **P < .01; ***P < .001. The data are shown as means ± SE of 3 independent experiments. The means and ranges of absolute concentrations were as follows: IFN-α, 74 312 pg/mL (48 118-92 759 pg/mL); IL-6, 3904 pg/mL (1970-6343 pg/mL).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/2/10.1182_blood-2010-05-284737/4/m_zh89991063850003.jpeg?Expires=1765894021&Signature=sRNtUlcFQRbAOLVo1GcDILgcqKnxLIcIeQKhmveHGrU2TcytRnSAOtA3PfRNUGiZfiiLamy5bU3W5RyJX1oL72JcgvcbU5CzzG9wSDTU08zMoEj70zBntV39NdXU7xtr6TsPV3yigIz-mizpXxy6SFfNN6C35LoV5A~ZGLTIQCRfSNtg2CpYmcv-rUblB97ORneVtxwakRu42Ig8Y1dv90z492JKo1WO1YCBROWO6RjJUkM~XSLM8s5nxL09m29AGamwep2XGK9RBQKLnbvuMqRxbjOY0PF1stAbsl9ynOyY0LJMS20kB073agAuFtvuGKuh-nzpvBFW0eoPw-rXyA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal