Abstract
Waldenström macroglobulinemia (WM) is a rare, lymphoplasmacytic lymphoma characterized by hypersecretion of immunoglobulin M (IgM) protein and tumor infiltration into the bone marrow and lymphatic tissue. Our understanding of the mechanisms driving the development and progression of WM is currently by the shortage of representative cell models available for study. We describe here the establishment of a new WM cell line, MWCL-1. Comprehensive genetic analyses have unequivocally confirmed a clonal relationship between this novel cell line and the founding tumor. MWCL-1 cells exhibit an immunophenotype consistent with a diverse, tumor clone composed of both small B lymphocytes and larger lymphoplasmacytic cells and plasma cells: CD3−, CD19+, CD20+, CD27+, CD38+, CD49D+, CD138+, cIgM+, and κ+. Cytogenetic studies identified a monoallelic deletion of 17p13 (TP53) in both the cell line and the primary tumor. Direct DNA resequencing of the remaining copy of TP53 revealed a missense mutation at exon 5 (V143A, GTG>GCG). In accordance with primary WM tumors, MWCL-1 cells retain the ability to secrete high amounts of IgM protein in the absence of an external stimulus. The genetic, immunophenotypic, and biologic data presented here confirm the validity of the MWCL-1 cell line as a representative model of WM.
Introduction
Waldenström macroglobulinemia (WM) is an indolent, non-Hodgkin lymphoma characterized by bone marrow infiltration and the overproduction of monoclonal immunoglobulin M protein (IgM).1 Accounting for 1% to 2% of hematologic malignancies, this rare lymphoma, which is often, if not always, preceded by an IgM monoclonal gammopathy of undetermined significance, is more common in males, with a median age at presentation of 63 years.2 The exact cause of WM is unknown, but multigenerational clustering and familial patterns suggest the possibility of an underlying genetic defect.3,4 However, cytogenetic analyses have yet to reveal a common abnormality specific to WM disease, as has been reported in other lymphoproliferative disorders.5,6 Whereas recent gene expression and proteomics studies have advanced our understanding of WM pathogenesis and identified potential therapeutic targets, WM remains incurable given current therapy, with a 5-year survival rate of 50%.7,8
WM is classified by the World Health Organization as a lymphoplasmacytic lymphoma, and bone marrow aspirates collected from patients with WM reveal a diverse tumor clone consisting of a spectrum of small B lymphocytes, plasma cells, and lymphoplasmacytoid cells.9 Evidence suggests that WM may arise from a postgerminal center, memory-like B cell that has developed the capacity to secrete IgM through either intrinsic or extrinsic stimuli. Thus, in addition to the positive expression of soluble immunoglobulin IgM and the pan B-cell antigens CD19 and CD20, many WM tumor cells also coexpress the markers of mature plasma cells as well, including CD38 and CD138.10,11
Clinical findings associated with this malignancy, including mucosal bleeding, retinopathies with visual disturbances, and neuropathies and paresthesias, frequently arise as a result of hyperviscosity syndrome, the development of which is directly related to the high serum levels of IgM.2,12 Despite its association with significant morbidity, the deregulation of IgM production in WM remains poorly understood. To date, exploratory studies to determine the pathogenesis of IgM dysregulation in WM have been limited mainly by the lack of a validated cell model that is both phenotypically and genetically consistent with the original primary tumor. The WSU-WM cell line was one of the first established from a WM tumor. However, WSU-WM cells maintain a karyotype and morphology more reminiscent of Burkitt lymphoma than WM; and because the cell line was never genetically authenticated against the primary tumor, the exact origin of these cells remains unclear.13 BCWM.1 cells correspond phenotypically to primary WM cells in their ability to secrete high levels of IgM, and this putative WM cell line has frequently been used as a representative model of this disease.14 Unfortunately, data confirming a clonal relationship between BCWM.1 cells and the primary tumor from which they were derived are limited. Although these cells remain a useful model for the study of IgM secretion and other B-cell phenomena, it is unclear whether or not BCWM.1 cells adequately represent WM.15 Thus, additional validated WM cell lines are prerequisite for increasing our understanding of this malignancy.
We report here on the establishment of a novel IgM-secreting, κ-immunoglobulin light chain–restricted, WM cell line, MWCL-1. Rigorous genetic testing over a 12-month period has unequivocally confirmed a clonal relationship between the primary WM tumor and MWCL-1 cells. In addition, MWCL-1 cells retain similar immunophenotypic and biologic properties of the initial tumor clone as well, including the cell surface expression of both B-cell and plasma cell markers as well as the ability to secrete high levels of IgM protein. Use of this validated cell model of WM disease will hopefully provide new insight into the development and progression of this rare lymphoma.
Methods
Case report and establishment of the cell line
Collection of bone marrow aspirates used in the establishment of WM cell lines was approved by the Mayo Foundation Institutional Review Board, and informed consent was obtained in accordance with the Declaration of Helsinki. The MWCL-1 cell line was established from the bone marrow aspirate of a 73-year-old male patient referred to the Mayo Clinic for management of his previously treated WM. He had been initially diagnosed with WM 10 years earlier and had received 3 previous courses of chemotherapy. At the time he was seen, he had evidence of retroperitoneal lymphadenopathy but no splenomegaly on computed tomography scan. His serum monoclonal IgM was 2.4 g/dL, and immunofixation confirmed a monoclonal IgM-κ. There was no evidence of hyperviscosity. His serum β2-microglobulin level was increased at 6.15 μg/mL (normal range, 0.7-1.8 μg/mL). The bone marrow aspirate from which the cell line was derived demonstrated a lymphoplasmacytic lymphoma composed of 50% of the bone marrow cellularity. By immunohistochemistry, the lymphoplasmacytic infiltrates contained CD20+ B cells and CD138+ plasma cells. Both the B cells and plasma cells were CD19+. Flow cytometry showed κ-light chain–restricted cells that were CD19+ and CD45+.
Unsorted bone marrow mononuclear cells were cultured in Iscove modified Dulbecco medium GlutaMAX (Invitrogen), supplemented with 50 U/mL penicillin G, 10 μg/mL gentamicin, 50 μg/mL streptomycin (Invitrogen), 10% heat-inactivated fetal calf serum (Atlanta Biologicals), 1 ng/mL interleukin-6 (IL-6; Peprotech), and 10 ng/mL insulin-like growth factor-I (Sigma-Aldrich). After 6 months, the cell line was cultured in RPMI 1640 (Invitrogen) supplemented with 50 U/mL penicillin G, 10 μg/mL streptomycin, 10% heat-inactivated fetal calf serum, and 1mM sodium pyruvate (Invitrogen).
Sequence analysis of IGHV
Total RNA was isolated using Trizol LS reagent (Invitrogen) as per the manufacturer's instructions, and RNA was reverse transcribed with Superscript III Reverse Transcriptase (Invitrogen). The cDNA (0.5 μg) was amplified (HotStar Taq Master Mix, QIAGEN) with a 5′ primer designed to hybridize to one of each of the 6 human variable H chain families (VH1 through VH6) and a 3′ antisense JH consensus primer that can hybridize to all 6 IGHV genes. Multiplex polymerase chain reaction (PCR) was performed using a PerkinElmer GeneAmp 9600 thermocycler (PerkinElmer Life and Analytical Sciences). All amplified PCR products were electrophoresed on a 1.5% agarose-TAE gel and visualized with ethidium bromide. For all amplifications, a β-actin reaction was also performed as an internal control. Band(s) were cut out and the DNA was extracted from the gel using MinElute Gel Extraction Kit (QIAGEN). The purified PCR product was sequenced with the 3′ primer at the Mayo DNA Sequencing Core Facility in Rochester, Minnesota. The sequences obtained were analyzed with a database of rearranged immunoglobulin genes, IMGT/V-QUEST (www.imgt.org/IMGT_vquest/share/textes), based on comparison of sequences for maximum nucleotide homology. The patient and cell line sequences were then aligned using Sequencher Version 4.5 (GeneCodes).
Immunofluorescence
Cytospin preparations of MWCL-1 cells on glass slides were prepared using a Thermo Shandon Cytospin 2 and stained with either FITC-conjugated anti-cIg antibody (Invitrogen) or FITC-conjugated anti-κ antibody (BD Biosciences PharMingen) before viewing by fluorescence microscopy (Olympus Provis AX70). Images were taken using an AxioCam ICc3 camera and Axionvison Version 4.8.1 software (Carl Zeiss MicroImaging). Images were edited with Adobe Photoshop CS2 Version 9.0.2 (Adobe Systems Inc).
Immunophenotypic analysis
MWCL-1 cells (0.5 × 106) were washed in 0.5% bovine serum albumin in phosphate-buffered saline and incubated with allophycocyanin-, phycoerythrin-, or FITC-conjugated antibodies for 30 minutes at 4°C. After washing and fixation, cells were analyzed using a FACSCalibur flow cytometer (BD Biosciences PharMingen) and CellQuest software (BD Biosciences). For qualitative assessment of expression, histograms of cells displaying the marker of interest were compared with those from cells stained with the respective isotype control. To characterize the intracellular immunoglobulin light chain, MWCL-1 cells were washed and resuspended in fixation medium (Fix and Perm Fixation Medium A, Invitrogen) for 15 minutes. After washing and permeabilization (Fix and Perm Permeabilization Medium B, Invitrogen), cells were incubated for 15 minutes in the presence of FITC-labeled anti-κ and anti-λ antibodies. Analysis was performed as for surface markers. Anti-transmembrane activator and calcium modulator and cyclophilin ligand interactor and the phycoerythrin-conjugated mouse IgG1 isotype control antibodies were purchased from R&D Systems. All other antibodies were from BD Biosciences PharMingen.
DNA fingerprinting
Genomic DNA was isolated from the original tumor and serial passages of the MWCL-1 cell line using Gentra Puregene Cell Lysis Buffer and Protein Precipitation Solution (QIAGEN). Extracted DNA was then analyzed for the presence of 34 single-nucleotide polymorphisms (SNPs) that have previously been reported to maximize the probability of obtaining distinct genotype profiles from different DNA samples.16 SNP array analysis was performed by the Mayo DNA Sequencing Core Facility in Rochester, Minnesota. The relatedness of the original tumor and the MWCL-1 cell line was determined by comparing the SNP profiles of the respective samples.
aCGH analysis
High-resolution array-based comparative genomic hybridization (aCGH) was performed with the Human Genome 244A microarray (Agilent Technologies). The digestion, labeling, and hybridization steps were done as previously described with minor modifications.17 Briefly, 1.2 mg of tumor and reference DNAs were separately digested with Bovine DNaseI (Ambion) for 12 minutes at room temperature. Random primers and exo-Klenow fragment (Invitrogen) were then used to differentially label tumor (Cy5) and reference (Cy3) genomic DNA samples (GE Healthcare). The labeled genomic reactions were cleaned up with Microcon YM-30 purification columns (Millipore) followed by hybridization at 65°C for 40 hours. A DNA Microarray Scanner (Agilent Technologies) was used to scan in microarray data, and feature extraction was performed with Agilent Feature Extraction Software Version 9.5. Log2 ratio data were imported and analyzed using Genomic Workbench Software Version 5.0.14 (Agilent Technologies).
FISH
The original archived bone marrow cell pellet (BM) and the MWCL-1 cell line (12-month passage) were stored at −70°C in methanol/glacial acetic acid (2:1) fixative. After a change of fixative, the fixed-cell pellet suspensions were manually dropped onto microscope slides and assessed by phase-contrast microscopy to verify appropriate cellularity. The BM and MWCL-1 samples were then subjected to standard fluorescence in situ hybridization (FISH) pretreatment, hybridization, and post-hybridization methods.18 The FISH probes used were p53 at 17p13 in orange with centromere 17 in green (Abbott Molecular) and c-MYB at 6q23 in orange with centromere 6 in green, which was made and validated at the Mayo Clinic. The BM and MWCL-1 cell line samples were analyzed and imaged using standard fluorescence microscopy methods.18 Microscopy was performed on a Leica DM5000 B microscope (Leica Microsystems) using a Leica HCX PL APO lens at 100×/1.40-0.7 oil objective. Cells were imaged in 10% DAPT/Vectashield anti-fade solution with an Applied Imaging 2912-5010/0000 camera and Genetix Cytovision Version 4.5.2 software (Genetix) used to acquire the digital images. Images were then edited using Adobe Photoshop CS2 (Adobe Systems Inc).
TP53 gene sequencing
Genomic DNA corresponding to exons 2 to 9 of TP53 was PCR amplified with GoTaq Hot Start Polymerase (Promega) and a Hybrid PCR Sprint Thermal Cycler (Thermo Scientific). The sequencing and amplification primers specific to TP53 exons 2 to 9 are listed in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The amplified PCR products were purified with QIAxcel DNA Large Fragment Kit (QIAGEN) before separation on a QIAxcel analyzer. Sequencing was performed using BigDye Terminator Version 1.1 Cycle Sequencing Kit (Applied Biosystems) and an ABI 3130XL Genetic Analyzer (Applied Biosciences) according to the manufacturer's protocol. Sequencher Version 4.5 software (GeneCodes) was used for TP53 sequence analysis.
Enzyme-linked immunosorbent assay
To quantitate the secretion of IgM, 0.5 × 106 cells were cultured in the presence or absence of specific cytokines in 24-well plates for 48 hours, after which the supernatants were harvested. IgM levels were determined via human IgM ELISA Kit following the manufacturer's protocol (Bethyl Laboratories). Cytokine concentrations were as follows: IL-2 (20 ng/mL), IL-6 and IL-10 (50 ng/mL), IL-12 (10 ng/mL), and B-cell activating factor (BAFF; 100 ng/mL).
Proliferation assay
To characterize cellular proliferation in response to individual cytokines and growth factors, MWCL-1 cells (2.5 × 104) were treated as indicated and cultured in 96-well plates. Cytokine concentrations used were as follows: IL-2, IL-4, IL-6, IL-8, IL-10, IL-12, epidermal growth factor, granulocyte colony-stimulating factor, IFN-α, and IFN-γ (10 ng/mL), BAFF (100 ng/mL), and CD40L (200 ng/mL). After 48 hours, the cells were pulsed with 1 μCi tritiated thymidine (3H-TdR, 10 Ci/mmol; PerkinElmer Life and Analytical Sciences) for an additional 24 hours and harvested onto glass fiber filter mats. 3H-TdR incorporation was determined on a PerkinElmer 1450 liquid scintillation counter (PerkinElmer Life and Analytical Sciences). Tritium accumulation in treated cells was then compared with accumulation in untreated controls.
Assessment of EBV infection
Evaluation of the Epstein-Barr virus (EBV) infection status of MWCL-1 cells was accomplished through PCR analysis of the EBV-encoded nuclear antigen 1 (EBNA1) gene as previously described.19 The presence of active viral shedding was determined by growing EBV− Karpas-422 cells in MWCL-1–conditioned media followed by PCR amplification of EBNA1 in the treated Karpas-422 cells.
Statistical analysis
All assays were performed in triplicate unless otherwise specified, and values are presented as the mean ± SD. Mean values were compared using the Student t test with differences between treatment groups considered statistically significant when the P value < .05 (Prism Version 5 software; GraphPad Software).
Results
Immunoglobulin analysis
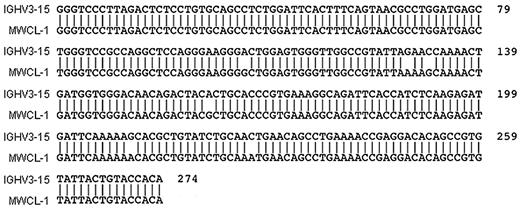
A comparison of VH gene usage in cells from the original bone marrow aspirate and the MWCL-1 cell line revealed identical VH sequences in both samples (Figure 1). VH3-15 was found to be the specific gene of use with 97.12% sequence identity (2.88% somatic hypermutation). In addition, flow cytometric and cytofluorescence immunophenotyping established monotypic κ-immunoglobulin light chain expression by both the primary tumor cells and MWCL-1 cells (Figure 2B). The presence of cytoplasmic IgM was also detectable by immunofluorescence in MWCL-1 cells.
Sequence alignment demonstrating homology between IgVH3–15 and the heavy chain genes of MWCL-1 cells and the primary tumor. PCR-amplified DNA was prepared and sequenced as discussed in “Sequence analysis of IGHV.” The sequence was compared with germline configurations using IMGT/V-QUEST software Version 3.2.17 on the IMGT website (www.imgt.cines.fr/IMGT_vquest/share/textes). Identical VH sequences were obtained for both MWCL-1 cells and the primary tumor, but this common sequence is only referred to as MWCL-1 in the figure.
Sequence alignment demonstrating homology between IgVH3–15 and the heavy chain genes of MWCL-1 cells and the primary tumor. PCR-amplified DNA was prepared and sequenced as discussed in “Sequence analysis of IGHV.” The sequence was compared with germline configurations using IMGT/V-QUEST software Version 3.2.17 on the IMGT website (www.imgt.cines.fr/IMGT_vquest/share/textes). Identical VH sequences were obtained for both MWCL-1 cells and the primary tumor, but this common sequence is only referred to as MWCL-1 in the figure.
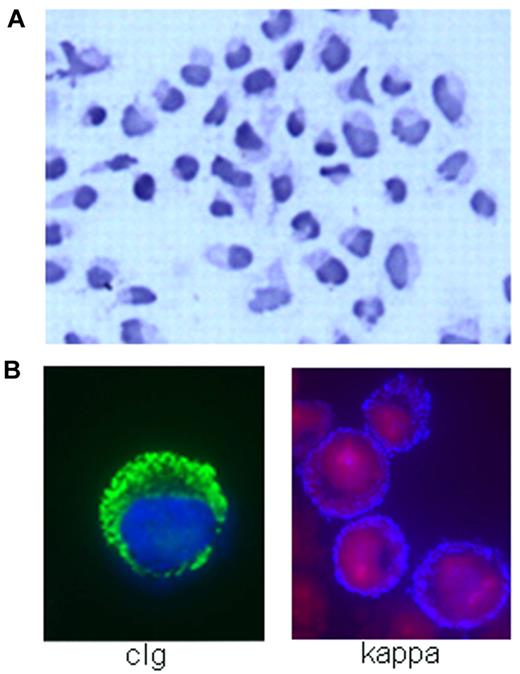
MWCL-1 cell line morphology. (A) Hematoxylin and eosin staining demonstrating the heterogeneity of the WM tumor clone represented in MWCL-1 cells, including B lymphocytes, lymphoplasmacytic cells, and mature plasma cells (original magnification ×200, Olympic Provus AX70 microscope). (B) Immunofluorescence showing the presence of κ-light chain on the extracellular surface of MWCL-1 cells and cytoplasmic IgM.
MWCL-1 cell line morphology. (A) Hematoxylin and eosin staining demonstrating the heterogeneity of the WM tumor clone represented in MWCL-1 cells, including B lymphocytes, lymphoplasmacytic cells, and mature plasma cells (original magnification ×200, Olympic Provus AX70 microscope). (B) Immunofluorescence showing the presence of κ-light chain on the extracellular surface of MWCL-1 cells and cytoplasmic IgM.
Characterization of cell morphology and immunophenotype
The morphology of MWCL-1 cells was consistent with that of a lymphoplasmacytic lymphoma, characterized by small B lymphocytes with densely staining nuclei as well as larger lymphoplasmacytic cells and mature plasma cells displaying irregular, eccentric nuclei and an abundance of cytoplasm (Figure 2A). Although the majority of cells were mononuclear, the presence of binucleated or multinucleated plasmacytic cells was not uncommon.
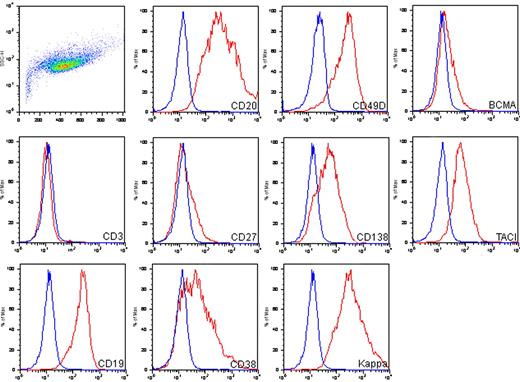
The immunophenotype of MWCL-1 cells, as determined by flow cytometry, was characterized by high expression levels of the B cell–associated antigens, CD19 and CD20, coexpressed with antigens associated with plasmacytic differentiation, including CD38, CD49d, and CD138 (Figure 3). In addition, CD19+ CD138− B lymphocytes and CD19− CD138+ plasma cells were present. Confirming the immunofluorescence data, expression of κ-immunoglobulin light chain was high as well. MWCL-1 cells were also positive for transmembrane activator and calcium modulator and cyclophilin ligand interactor and BCMA, the receptors for tumor necrosis factor family members BAFF and a proliferation-inducing agent, both of which are highly involved in the regulation of B-cell malignancies, including WM.20 CD27 was dimly expressed on MWCL-1 cells, and expression of the T-cell marker CD3 was negative.
DNA fingerprinting
The authenticity of the MWCL-1 cell line was determined through the use of a validated SNP panel identification assay, which allows for the detection of unique DNA fingerprints through the genotyping of 34 predetermined SNPs (data not shown).16 The original WM tumor and the MWCL-1 cell line shared 100% of genotype calls (all 34 SNPs), confirming the parentage of the MWCL-1 cell line. To monitor genetic stability of the MWCL-1 cell line over time, serial passages of MWCL-1 cells were genotyped and compared with the primary tumor fingerprint. The SNP profile of high passage number MWCL-1 cells remained identical to that of the founding tumor cells.
aCGH analysis
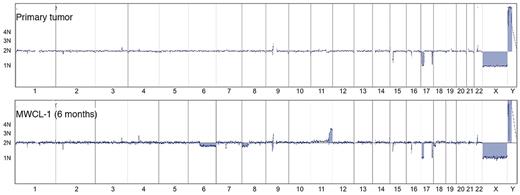
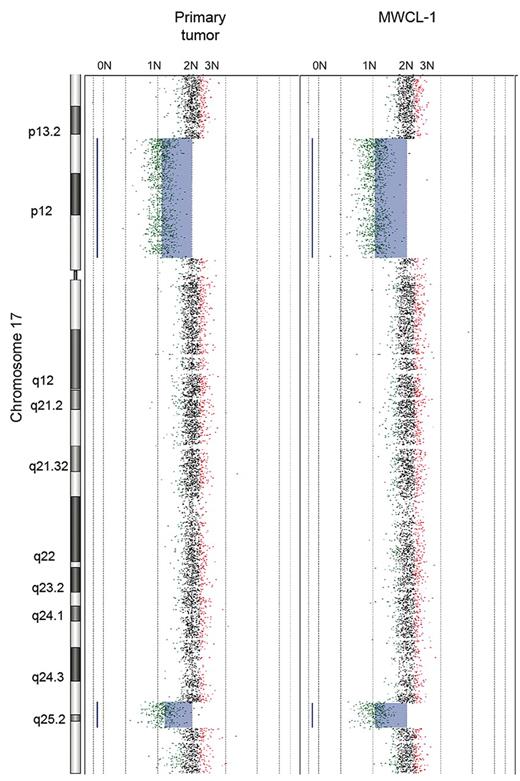
The aCGH profile clearly confirms that the MWCL-1 cells are genetically related with the primary patient sample (Figure 4). The karyotype of the parental cells was characterized by deletions in 2p21, 17p11-p13.2, and 17q25.1-q25.3. These losses, sharing the same breakpoints, were also detected in the MWCL-1 cells at 6 months and 12 months, thus confirming that MWCL-1 cells share common copy number abnormalities (CNAs) with the original founding tumor. A more detailed view of chromosome 17 is shown in Figure 5. In addition, chromosomal losses at 6q12.3-qter and 8p and 2 extra copies of 11q24.1-qter were found in MWCL-1 cells at 6 months. At this time, the 6q12.3 deletion was present in a small subclone of cells, but at later passages, this abnormality was absent or below the limit of sensitivity of both aCGH and FISH assays (Figures 5, 6A). Similarly, the 6q deletion was undetectable in the original tumor based on the sensitivity limits of aCGH, and no additional sample was available to validate the data through FISH or another approach.
Immunophenotypic characterization of the MWCL-1 cell line. FITC-, allophycocyanin-, and phycoerythrin-conjugated antibodies were used for phenotyping of the MWCL-1 cell line by flow cytometry. Blue peaks represents isotype staining; and red peaks, antigen-specific staining.
Immunophenotypic characterization of the MWCL-1 cell line. FITC-, allophycocyanin-, and phycoerythrin-conjugated antibodies were used for phenotyping of the MWCL-1 cell line by flow cytometry. Blue peaks represents isotype staining; and red peaks, antigen-specific staining.
Whole-genome aCGH profiles. Comparative chromosomal CNAs in the original WM patient sample and the established MWCL-1 cell line at a 6-month passage. Note the common CNAs on chromosome 17 between all samples. Chromosomes 1 through 22, X, and Y, from p to q arm, are plotted on the x-axis from left to right. The predicted number of copies of each gene is indicated to the left of each panel.
Whole-genome aCGH profiles. Comparative chromosomal CNAs in the original WM patient sample and the established MWCL-1 cell line at a 6-month passage. Note the common CNAs on chromosome 17 between all samples. Chromosomes 1 through 22, X, and Y, from p to q arm, are plotted on the x-axis from left to right. The predicted number of copies of each gene is indicated to the left of each panel.
Primary WM tumor and MWCL-1 cell line share common chromosome CNAs. Whole chromosome plot of chromosome 17 is shown for the original WM patient sample and the established MWCL-1 cell line at 6 months, illustrating the interstitial losses in 17p13-p13.2 and 17q25.1-q25.3. The predicted number of copies of each gene is indicated on the top of each panel.
Primary WM tumor and MWCL-1 cell line share common chromosome CNAs. Whole chromosome plot of chromosome 17 is shown for the original WM patient sample and the established MWCL-1 cell line at 6 months, illustrating the interstitial losses in 17p13-p13.2 and 17q25.1-q25.3. The predicted number of copies of each gene is indicated on the top of each panel.
TP53 analysis
aCGH and FISH analysis of the MWCL-1 cell line detected a monoallelic loss of chromosome 17p13, including TP53 (Figure 6B). To further characterize the status of TP53, direct DNA resequencing was performed. Coding exons 2 to 10 of TP53 were sequenced from genomic DNA isolated from MWCL-1 cells at 12 months, and a missense mutation at exon 5 (V143A, GTG > GCG) was identified. The presence of this same mutation was also confirmed in the original tumor biopsy, thus confirming that both copies of TP53 were affected in the original patient sample.
MWCL-1 cells retain both copies of chromosome 6q but exhibit monoallelic expression of mutated TP53. (A) Representative FISH image of MWCL-1 cells at a 12-month passage confirming the absence of a 6q deletion. Two hybridization signals are observed for both the probe specific to the centromere of chromosome 6 (green signal), and the locus-specific, c-MYB probe (red signal). (B) Representative FISH image of MWCL-1 cells at a 12-month passage demonstrating monoallelic loss of TP53. Two hybridization signals are observed for the probe specific to chromosome 17 (green signal), but only one signal is observed for the locus-specific, TP53 probe (red signal).
MWCL-1 cells retain both copies of chromosome 6q but exhibit monoallelic expression of mutated TP53. (A) Representative FISH image of MWCL-1 cells at a 12-month passage confirming the absence of a 6q deletion. Two hybridization signals are observed for both the probe specific to the centromere of chromosome 6 (green signal), and the locus-specific, c-MYB probe (red signal). (B) Representative FISH image of MWCL-1 cells at a 12-month passage demonstrating monoallelic loss of TP53. Two hybridization signals are observed for the probe specific to chromosome 17 (green signal), but only one signal is observed for the locus-specific, TP53 probe (red signal).
Biologic characterization of MWCL-1 cells
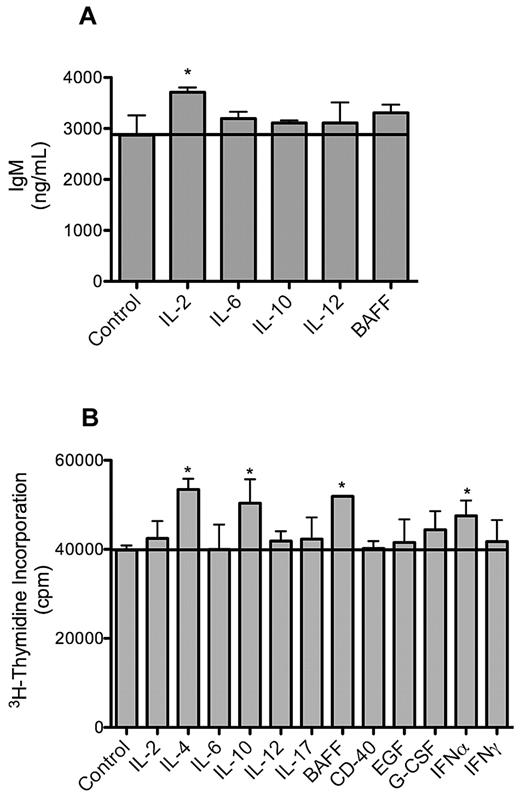
Consistent with primary WM cells, the MWCL-1 cell line retained the ability to secrete IgM in the absence of an external stimulus over the entire 12-month study period. IgM secretion in MWCL-1 cells was enhanced in the presence of cytokines with known effects on immunoglobulin production and secretion. Namely, treatment with IL-2 produced the most significant increase in IgM secretion (P < .001, Figure 7).
IgM secretion and proliferation of MWCL-1 cells in response to cytokines. MWCL-1 cells were cultured with the indicated cytokines for 72 hours, at which time IgM secretion was quantified by enzyme-linked immunosorbent assay (A), and DNA synthesis measured by 3H-TdR incorporation (B). Bars represent the mean of triplicate experiments ± SD. *Value significantly higher than in the respective untreated control cells (P < .05).
IgM secretion and proliferation of MWCL-1 cells in response to cytokines. MWCL-1 cells were cultured with the indicated cytokines for 72 hours, at which time IgM secretion was quantified by enzyme-linked immunosorbent assay (A), and DNA synthesis measured by 3H-TdR incorporation (B). Bars represent the mean of triplicate experiments ± SD. *Value significantly higher than in the respective untreated control cells (P < .05).
Similarly, proliferation in response to cytokines was assessed in MWCL-1 cells as well. Cytokine-induced cell growth in this cell line was more modest than has been previously reported for other non-WM plasma cell lines, but an increase in the proliferation was still observed after treatment with BAFF, IL-4, IL-10, and interferon-α (P < .001, Figure 6).21,22
Lastly, genomic DNA isolated from MWCL-1 cells tested positive for the EBV gene product, EBNA1. To determine whether the virus was actively replicating and being shed by MWCL-1 cells, EBV− Karpas-422 cells were treated with MWCL-1–conditioned media for 96 hours. EBNA1 was not detectable in the Karpas-422 cells at the end of the treatment period.
Discussion
A major factor prohibiting the study of WM pathogenesis is the lack of a model cell line possessing both a phenotype and genotype with overall similarity to the original tumor. In this study, we describe the development and extensive characterization of a novel WM cell line, MWCL-1. Unlike with other putative WM cell lines, multiple, highly sensitive analyses were used to establish an unequivocal genetic link between MWCL-1 cells and the founding tumor. Heavy chain sequence analysis yielded common clone-specific VH gene usage in both the primary WM tumor and the MWCL-1 cell line, providing direct evidence of the clonal relationship between the 2 samples. DNA fingerprinting of both the cell line and the original tumor revealed identical genomic profiles as well, further confirming the maintenance of the tumor genotype throughout the establishment of the MWCL-1 cell line. In addition, the potential for genetic drift or contamination of cell lines in long-term culture has been addressed by routine DNA fingerprinting of the MWCL-1 cells over a period of 1 year. Again, a consistency of genotype between the primary tumor cells and the MWCL-1 cell line has been maintained, regardless of passage number.
Moreover, array CGH has provided detailed chromosomal maps by which the genomes of the MWCL-1 cell line and the parent tumor were compared, and the data again strongly demonstrate the relatedness of the samples. The karyotypes of both the primary tumor and the MWCL-1 cells exhibit regional losses in 2p21, 17p11-p13.2 (includes TP53), and 17q25.1-17q25.3. Deletions to 17p are not unique to MWCL-1 cells, having been observed in nearly all subtypes of non-Hodgkin lymphoma.23 Loss of this chromosome is predictive for a poorer prognosis in many low-grade lymphomas, with further evidence associating abnormalities of chromosome 17 in lymphoma with lower survival, regardless of histologic grade.23-26 Although relatively uncommon, deletions in 17p13.1 and the subsequent loss of p53 have been identified previously in patients with WM as well, this karyotype having been associated with higher clonal involvement of the bone marrow.5,17,26
The status of p53 in the MWCL-1 cell line was further characterized by FISH and direct gene resequencing approaches. FISH analysis revealed a monoallelic deletion of p53 in 100% of cells. As has been commonly noted in various human malignancies, the loss of one p53 allele in the MWCL-1 cell line was coupled with a missense mutation in the other allele.27 The specific mutation identified in MWCL-1 cells was consistent with that detected in genomic DNA isolated from the original patient sample and involves the substitution of a valine for an alanine at amino acid 143. The resulting mutated protein, p53V143A, is temperature-sensitive, possessing no transcriptional activity at 37°C.28 Although the activation of genes involved in growth arrest and DNA repair has been reported at lower temperatures, the location of the mutation within the DNA binding site still renders p53V143A defective in inducing apoptosis. Although the role of p53 in the pathogenesis of other malignancies has been thoroughly evaluated, the exact relationship between p53 activity and the pathogenesis of WM is not well understood. In a recent study, 8.1% of WM patients were reported to exhibit a p53 deletion, and loss of this tumor suppressor gene was significantly associated with a shorter time to progression.6 Thus, it seems likely that, as with other malignancies, the absence of p53 may be one of many factors driving the development and progression of WM.
Aside from the 17p13 deletion, aCGH analysis did not detect other chromosomal abnormalities previously identified in WM, consistent with the lack of an original cytogenetic signature in this disease. One of the most commonly observed abnormalities in WM is deletion of 6q, with a prevalence ranging from 22% to 54%.17,29 Although the loss of 6q is not unique to WM and has not been associated with overall survival, initial data suggest that this abnormality may be predictive of a longer treatment response.6 Interestingly, the loss of 6q was detected in a small subclone of MWCL-1 cells at the 6-month passage. However, this abnormality was not detectable in the initial patient sample or in later passages, suggesting that the proliferation of cells possessing this particular deletion was suppressed by a more dominant clone.
Phenotypically, MWCL-1 cells are best defined as a κ-immunoglobulin light chain-restricted lymphoplasmacytic cell line, consistent with the parental tumor cells. As is characteristic of WM tumors, MWCL-1 cells retain the ability to secrete intact IgM, although only treatment with IL-2 was able to significantly enhance IgM secretion. These data are consistent with previous studies performed in primary WM cells, where it was observed that a combination of cytokines is required to significantly increase the secretion of IgM.20 Interestingly, the putative WM cell line, BCWM.1, secretes IgM at a much lower baseline rate than MWCL-1 cells and primary cells yet remains more responsive to stimulation by individual cytokines (data not shown). Thus, it is possible that the biologic processes and signaling pathways regulating IgM secretion in MWCL-1 cells may be more similar to those used by primary WM cells than BCWM.1 cells, providing further support for the use of the MWCL-1 cell line as an experimental model of WM disease.
Lastly, PCR analysis revealed the MWCL-1 cell line to be EBV positive, although further studies demonstrated that the cells were not actively shedding EBV virus. One concern with the development of WM disease models is whether or not tumor-derived cell lines are actually representative of the original WM tumor clone itself or are merely bystander B cells that have been immortalized by EBV.15 EBV-transformed B lymphocytes often outgrow neoplastic cells; and because they possess similar morphologic and biologic characteristics as their malignant counterparts, it is often difficult to determine the exact derivation of a particular cell line without extensive genetic testing.30 In characterizing the EBV+ MWCL-1 cell line, we have avoided the ambiguity associated with the origins of previous EBV+ WM cell lines by undertaking not only comprehensive genetic testing, but also immunophenotypic and biologic testing.
The results presented here confirm the relatedness of MWCL-1 cells and the primary tumor from which they were originally isolated. In addition, this extensive characterization has provided substantive new insights into the unique genotype and phenotype of WM cells. Authenticated cell lines, such as MWCL-1, will prove essential in future studies into the pathogenesis of this incurable disease.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Lori A. Frederick and Ryan A. Knudson from the Divisions of Hematopathology and Cytogenetics, respectively, at the Mayo Clinic, Rochester, MN, for their assistance with the TP53 analysis.
This work was supported in part by the Leukemia & Lymphoma Society, the International Waldenström Macroglobulinemia Foundation, and the Predolin Foundation.
Authorship
Contribution: A.J.N. and S.M.A. designed research; L.S.H., D.M.G., E.B., M.K.M., T.L.P.T., and S.C.Z. performed research and collected data; L.S.H., A.J.N., R.P.K., R.F., T.E.W., W.G.M., M.A.G., and S.M.A. analyzed and interpreted data; and L.S.H. and S.M.A. wrote the paper with input from all authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Stephen M. Ansell, Division of Hematology, Mayo Clinic, 200 First St SW, Rochester, MN 55905; e-mail: ansell.stephen@mayo.edu.