Abstract
Immunoglobulin (Ig) gene rearrangements are used to define clonality of suspected B-lineage malignancy in tissue samples. To determine whether such rearrangements could be identified in plasma, we screened plasma from 14 consecutive patients with AIDS-related lymphoma with multiplex Ig primers. Clonally rearranged Ig DNA was detected in plasma from 7 of 14 patients. Patients in whom clonal Ig DNA remained detectable after combination chemotherapy died with lymphoma. Tumor was available from 1 patient, and the IgH amplification products from plasma and tumor were sequenced and confirmed to be identical. Ig DNA rearrangements in plasma may be useful as a lymphoma-specific tumor marker, and failure to clear clonal Ig DNA may identify patients at high risk for failure of standard therapy.
Introduction
Molecular diagnostic tests are widely used to detect clone-specific immunoglobulin (Ig) rearrangements. Circulating nucleic acids have been explored as tumor markers in other settings. Preliminary studies have defined the value of clonal Ig DNA in the serum or plasma as a potential marker in lymphoma patients.1,2 Since these early reports, polymerase chain reaction technology has evolved. Capillary electrophoresis and the use of standardized primer sets have enhanced reliability and sensitivity in DNA extracted from diagnostic biopsy specimens.3-6
A higher incidence of non-Hodgkin lymphoma is seen in persons infected with HIV compared with the general population, even in the era of effective antiretroviral therapy.7 The signs and symptoms of lymphoma, such as lymphadenopathy and fever, may overlap with those of HIV disease progression and associated opportunistic infections.8 Extranodal presentations of lymphoma are more common in patients with HIV. Monitoring clinical responses to therapy in patients who have AIDS-related lymphoma (ARL) also presents special challenges. Fluorodeoxyglucose avidity, as measured by positron emission tomography, may reflect HIV infection per se, immune reconstitution after antiretroviral therapy, or opportunistic infection.9,10 With these complexities in mind, we sought to explore the possible utility of detection of clonally rearranged Ig DNA in plasma in ARL.
Methods
Plasma and peripheral blood mononuclear cell specimens from healthy blood donors, patients with ARL, and patients with AIDS Kaposi sarcoma were obtained with informed consent in accordance with the Declaration of Helsinki after approval by institutional review boards of all participating institutions. Plasma was separated from peripheral blood collected in standard ethylenediaminetetraacetic acid or acid citrate dextrose tubes and stored at −80°C. DNA was extracted from 500 μL of plasma with QIAamp DNA blood mini-kit (QIAGEN). Fluorochrome-labeled, standardized multiplex primers (BIOMED2) targeting IgH Fr1-JH, Fr2-JH, Fr3-JH, and DH-JH, and IgK at Vk-Jk and Vk-Kde (InVivoScribe Technologies) were used. Polymerase chain reaction products were analyzed with an ABI 3100 with tetramethylrhodamine size standards (Applied Biosystems). To classify a specimen as clonal, we required that the peak height of the spike be greater than 2-fold over background. Assays were performed in duplicate.
Results and discussion
We evaluated multiplex primers in pretreatment plasma from 14 consecutive ARL patients. Among these, 10 had a diagnosis of diffuse large B-cell lymphoma, 3 had large B-cell lymphoma not otherwise specified, and one had primary effusion lymphoma. Clonally rearranged Ig DNA was detected in plasma from 7, including 6 IgH and 4 IgK rearrangements (Table 1). In 1 case (patient 4), tumor tissue was available and the identity of the Ig rearrangements in plasma and tumor were confirmed (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Clonal peaks were not identified in corresponding peripheral blood mononuclear cells. Similarly, clonal peaks were not identified in the plasma from 10 patients with AIDS KS but without lymphoma and from 10 healthy subjects.
Profile demonstrating the location of rearrangements in patients with detectable clonal Ig DNA
| Patient no. . | IGH primers . | IGK primers . | |||||
|---|---|---|---|---|---|---|---|
| Fr1 . | Fr2 . | Fr3 . | DH1-6 . | DH7 . | Vk . | Kde . | |
| 1 | + | + | |||||
| 2 | + | + | |||||
| 4 | + | ||||||
| 6* | + | + | |||||
| 9 | + | + | + | + | |||
| 11 | + | ||||||
| 13 | + | ||||||
| Patient no. . | IGH primers . | IGK primers . | |||||
|---|---|---|---|---|---|---|---|
| Fr1 . | Fr2 . | Fr3 . | DH1-6 . | DH7 . | Vk . | Kde . | |
| 1 | + | + | |||||
| 2 | + | + | |||||
| 4 | + | ||||||
| 6* | + | + | |||||
| 9 | + | + | + | + | |||
| 11 | + | ||||||
| 13 | + | ||||||
Fr indicates framework region; DH, diversity region; Vk, variable κ region; Kde, κ-deleting element (ie, λ restricted); +, present.
Fr3 screened alone for IgH.
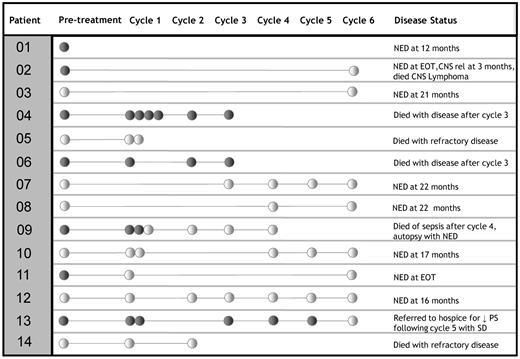
Serial specimens were available from 13 patients (Figure 1). Two patients (patients 4 and 6) had persistent clonal spikes and died of refractory disease during chemotherapy. Two patients (patients 9 and 11) had spikes that disappeared within 2 weeks of the initiation of chemotherapy and remained absent throughout their treatments. One achieved clinical remission (patient 11), whereas the other (patient 9) died with sepsis after cycle 4, and there was no evidence of lymphoma at autopsy. A patient (patient 13) with primary effusion lymphoma had detectable clonal Ig DNA at baseline through cycle 5 of treatment and then disappearance of clonal Ig DNA at cycle 6. Declining performance status led to transition to palliative care, and no further specimens were obtained. Patient 2 had disappearance of a clonal spike and achieved complete remission. He subsequently developed central nervous system lymphoma. A new, different plasma spike appeared. With the initiation of treatment, this spike was no longer detectable. No further specimens were obtained, and the patient died in hospice. Among 7 patients with no detectable clonal Ig DNA in the plasma at baseline, clonal Ig DNA remained undetectable in 5 throughout their treatment, and these patients remain in remission at 16, 17, 21, 22, and 22 months from the time of diagnosis. Two patients without clonal Ig DNA at baseline died of refractory disease.
Serial specimens from ARL patients. Positive specimen (clonal Ig DNA detected) is indicated by ●; and negative specimen (no clonal Ig DNA detected) by ○. NED indicates no evidence of disease; EOT, end of treatment; rel, relapse; PS, performance status; and SD, stable disease.
Serial specimens from ARL patients. Positive specimen (clonal Ig DNA detected) is indicated by ●; and negative specimen (no clonal Ig DNA detected) by ○. NED indicates no evidence of disease; EOT, end of treatment; rel, relapse; PS, performance status; and SD, stable disease.
We detected clonally rearranged Ig DNA in half of the patients studied. Inability to detect clonal Ig DNA in the remaining patients may reflect somatic hypermutation or other alterations in the Ig sequences that preclude the primers used from amplifying Ig DNA or may reflect low tumor cell turnover. In healthy persons, and in patients with HIV infection but without lymphoma, clonal Ig DNA was not detected. Because a B cell can only release its uniquely rearranged Ig DNA once and the half-life of DNA in plasma is measured in minutes or hours,11,12 the mass of clonal tumor cells that generate circulating DNA must continue to proliferate and must reflect a substantial tumor burden. This contrasts with detection of monoclonal Ig protein spikes that may reflect the presence of long-lived plasma cells that may secrete protein over years. Thus, it is not surprising that clonally rearranged Ig DNA was not detected in healthy subjects or AIDS patients without lymphoma.
We note that we found 2 or more rearrangements in only 4 of our 7 patients. However, the substrate for our tests (plasma DNA) is, on average, quite small in size and is therefore probably undetectable using several of the BIOMED-2 primer sets, akin to DNA from formalin-fixed tissue (as discussed in the original BIOMED-2 report).3 In a follow-up report, these authors detailed that 18% (20 of 109) of diffuse large B-cell lymphoma tissue specimens had a single rearrangement using both the IgH and IgK primer sets.6 Taking into account the unique limitation of the plasma DNA substrate, this is not significantly different from the 3 of 7 with one rearrangement in our study. In addition, the patient (patient 4) that confirmed the tumor derivation of plasma clonal Ig rearrangements had only one clonal band in the plasma, further supporting the notion that a single rearrangement may be sufficient evidence for clonality.
Detecting clonal Ig DNA may be especially useful in ARL given that the great majority of ARLs are of B-cell origin and commonly display high proliferative rates. Detection of clonal Ig DNA might ultimately serve to prompt an earlier diagnostic biopsy in some patients with nonspecific symptoms or signs. In addition, Ig DNA detection studies showing persistence of DNA after the initiation of therapy might guide a response-adapted or individualized therapeutic or palliative approach. In each of the 2 cases in this study in which Ig DNA persisted after the initiation of chemotherapy, the patients died as the result of progressive tumor growth. As will be appreciated from Figure 1, the availability of follow-up specimens was uneven, reflecting that the main focus of specimen collection related to assessment of Epstein-Barr virus DNA rather than Ig DNA and the Ig DNA studies were only undertaken when excess specimen was available. Further studies are needed to establish the predictive value and clinical utility of detection of clonal Ig DNA in plasma before a diagnosis of lymphoma, or after the initiation of therapy. The continued technologic advances in detection and characterization of DNA, combined with the unique characteristics of Ig DNA, suggest that such investigations may be fruitful in many high-grade B-cell malignancies and not just in HIV-associated lymphoma.
Presented in abstract form at the 49th Annual Meeting of the American Society of Hematology, Atlanta, GA, December 8-11, 2007.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Institutes of Health (National Research Service Award training grant 5T32 CA09071), the Johns Hopkins Lymphoma SPORE (P50 CA968888), and the AIDS Malignancy Consortium (U01CA121947).
National Institutes of Health
Authorship
Contribution: L.G., N.D.W.-J., and R.F.A. conceived the studies; L.G. and C.D.G. performed experiments; C.D.G., V.M.L., O.M.-M., N.D.W.-J., J.L., and R.F.A. analyzed results; and N.D.W.-J. and R.F.A. wrote the paper.
Conflict-of-interest disclosure: C.D.G. is the cofounder and co-owner of OncoMedx and is also a member of the Board of Directors and a manager of the company. OncoMedx has licensed several patents from Pennsylvania State University. As a coinventor of those patents, C.D.G. is entitled to a share of the licensing income. The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its conflict-of-interest policies. The remaining authors declare no competing financial interests.
Correspondence: Nina D. Wagner-Johnston, Washington University School of Medicine, Division of Oncology, 660 S Euclid Ave, Box 8056, St Louis, MO 63110; e-mail: nwagner@dom.wustl.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal