The urokinase plasminogen activator receptor (uPAR) has emerged as a potential regulator of cell adhesion, cell migration, proliferation, differentiation, and cell survival in multiple physiologic and pathologic contexts. The urokinase plasminogen activator (uPA) was the first identified ligand for uPAR, but elucidation of the specific functions of the uPA-uPAR interaction in vivo has been difficult because uPA has important physiologic functions that are independent of binding to uPAR and because uPAR engages multiple ligands. Here, we developed a new mouse strain (PlauGFDhu/GFDhu) in which the interaction between endogenous uPA and uPAR is selectively abrogated, whereas other functions of both the protease and its receptor are retained. Specifically, we introduced 4 amino acid substitutions into the growth factor domain (GFD) of uPA that abrogate uPAR binding while preserving the overall structure of the domain. Analysis of PlauGFDhu/GFDhu mice revealed an unanticipated role of the uPA-uPAR interaction in suppressing inflammation secondary to fibrin deposition. In contrast, leukocyte recruitment and tissue regeneration were unaffected by the loss of uPA binding to uPAR. This study identifies a principal in vivo role of the uPA-uPAR interaction in cell-associated fibrinolysis critical for suppression of fibrin accumulation and fibrin-associated inflammation and provides a valuable model for further exploration of this multifunctional receptor.
Introduction
The urokinase plasminogen activator receptor (uPAR) is a glycosylphosphatidylinositol-anchored glycoprotein that is expressed by many hematopoietic, connective tissues, and epithelial cells in particular during physiologic and pathologic tissue remodeling processes.1 uPAR has been reported to interact with a remarkable array of proteins, including extracellular matrix molecules, cell adhesion receptors, G-protein–coupled receptors, tyrosine kinase receptors, and internalization receptors. In this capacity, uPAR is reported to regulate cell adhesion, cell migration, proliferation, chemotaxis, differentiation, cell survival, cellular senescence, oncogenesis, invasion, angiogenesis, efferocytosis, and more. uPAR was originally identified as a high-affinity cellular binding site for urokinase plasminogen activator (uPA), a principal activator of plasminogen. The requirement of uPA binding to uPAR for the plethora of these uPAR-dependent cellular processes is frequently dependent on the specific experimental ex vivo context in which these processes are being studied, and the contribution of uPA binding per se to uPAR in regulating the activities of the receptor in vivo in most cases is not well established (reviewed in Smith and Marshall2 and Blasi and Carmeliet3 ; see also supplemental References 1-22, available on the Blood website; see the Supplemental Materials link at the top of the online article).
Genetics has proven to be a powerful tool for the functional dissection of ligand-receptor interactions. Current animal models to study the uPA-uPAR interaction include uPA-null mice,4 uPAR-null mice,5,6 and mice with tissue-specific overexpression of uPA and uPAR.7 Each of these animal models has proved exceptionally useful in the exploration of the uPA-uPAR system.8 However, the wholesale ablation or overexpression of either uPA or uPAR is not ideal for the dissection of the specific physiologic and pathologic functions of the uPA-uPAR interaction, because uPAR engages multiple ligands besides uPA, and because uPA is critical for tissue homeostasis and tissue repair independently of binding to uPAR9,,,–13 (see “Results”).
Therefore, we generated a novel mouse strain in which the uPA-uPAR interaction is selectively abrogated, whereas all other functions of the protease and its cellular receptor are retained. We use the novel mouse strain to delineate the requirement of uPA binding to uPAR for long-term health, inflammation, tissue homeostasis, fibrin surveillance, and wound repair, and we identify a novel role of the uPA-uPAR interaction in suppressing chronic inflammation secondary to exacerbated fibrin deposition.
Methods
Generation of PlauGFDhu/GFDhu mice
Gene targeting was performed by homologous recombination in Bruce4 embryonic stem (ES) cells derived from C57BL/6 mice.14 The targeting vector was constructed from 2 Plau genomic DNA fragments: A 6.6 Kb 5′ homology arm with the knock-in mutations incorporated at the 3′ end and a 2.7 kb 3′ homology arm. These 2 DNA fragments were obtained by polymerase chain reaction (PCR) from the C57BL/6 mouse genome, confirmed by DNA sequencing, and cloned into pLOz, a plasmid from Ozgene Pty containing a PKG-Neo expression cassette flanked by 2 loxP sites, to create the uPA knock-in targeting vector. Electroporation, blastocyst injections, screening, and initial mouse-breeding work were performed by Ozgene Pty under contract from the National Institute of Allergy and Infectious Diseases, National Institutes of Health. The neo cassette was removed by crossing germline chimeric mice to the Oz_Cre deletor strain (Ozgene Pty).
Mouse husbandry
The generation of Plau and Plaur gene-targeted mice has been described previously.4,6,9,15 The mice were backcrossed at least 7 times to C57BL/6J mice. The mice were housed in an Association for Assessment and Accreditation of Laboratory Animal Care–certified facility, and the study was carried out in accordance with institutional guidelines with approval from the National Institutes of Health. The genotype of the animals was determined by PCR with use of DNA derived from ear biopsies at weaning, and the genotype was confirmed for each mouse at the termination of the experiment. The targeted and endogenous Plau and Plaur alleles were detected as described previously.9 uPA knock-in mice were genotyped with use of the following primer pairs: PlauGFDhu/GFDhu allele: 5′- AACAAGTACTTCTCCAACATTCACTGG-3′ and 5′- GGGACTGGGGGTACAGATGGC-3′. Plau+/+ allele: 5′- TACAAGTACTTCTCCAGAATTCGCCGA-3′ and 5′- GGGACTGGGGGTACAGATGGC-3′.
Quantitative real-time PCR analysis
Total RNA from mouse kidney was prepared by extraction in Trizol reagent (Gibco-Invitrogen), as recommended by the manufacturers. First-strand cDNA synthesis was performed with use of Oligo(dT) primers with the RETROscript reverse transcription kit (Applied Biosystems/Ambion). The Bio-Rad iCycler, Gene Expression Analysis Software, and IQ SYBR Green Supermix (Bio-Rad Laboratories) were used for real-time reverse transcription (RT)–PCR in accordance with the manufacturer's directions, with use of a primer complementary to a sequence in Plau exon 8, 5′-GAAGTTTGAGGTGGAGCAGC-3′ and Plau exon 9, 5′- CCCGTGCTGGTACGTATCTT-3′ (annealing temperature 59°C, denaturation temperature 94°C, 40 cycles). Plau expression levels were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels in each sample. GAPDH mRNA was amplified with the primers 5′- GTGAAGCAGGCATCTGAGG -3′ and 5′- CATCGAAGGTGGAAGAGTGG-3′.
Western blot of uPA
Samples of pooled void urine from defined mouse genotypes were subjected to SDS-PAGE on a 12% (wt/wt) Bis-Tris polyacrylamide gel (NuPAGE; Invitrogen) under nonreducing conditions. The separated proteins were transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore) by electroblotting, and additional binding sites were blocked with 2% (wt/vol) bovine serum albumin (BSA) in 0.05% (vol/vol) Tween 20 in phosphate-buffered saline (PBS). Mouse uPA was detected by incubation overnight at 4°C with 2 μg/mL purified polyclonal rabbit anti–mouse uPA antibody (prepared in-house), followed by a peroxidase-labeled swine anti–rabbit immunoglobulin G (IgG; DAKO).
Ligand blotting
Recombinant mouse uPAR or human uPAR (0.5 μg)16 was separated by SDS-PAGE and transferred to PVDF membranes as described in “Western blot of uPA.” These membranes were incubated overnight at 4°C with either 1nM purified recombinant human or murine pro-uPA16,17 or with void urine collected from defined genotypes and diluted 20-fold in PBS containing 0.05% (vol/vol) Tween 20. The receptor-bound uPA was detected by incubation with 2 μg/mL of purified polyclonal rabbit anti–mouse or rabbit anti–human uPA following the protocol outlined in “Western blot of uPA.”
uPA-activated anthrax toxin intoxication
PlauGFDhu/GFDhu mice and littermate Plau+/+ mice were housed together and injected intraperitoneally with 200 μg of PrAg-U2 and 10 μL of FP59 in PBS.18 The mice were monitored every 12 hours for signs of intoxication by an investigator who was blinded to mouse genotype. The mice were humanely killed when they showed signs of pain and distress.
Assessment of long-term health
Groups of 8 to 10 female and male mice of the various genotypes were housed together and monitored weekly for outward appearance and survival. At age 1 year, mice were injected with 100 units of heparin in 100 mL of PBS, before perfusion with PBS, as described.19 Tissues were then snap-frozen or processed into paraffin.
Histologic examination of tissues
Tissues were fixed for 24 hours in Z-fix (Anatech), embedded into paraffin, sectioned, stained with hematoxylin & eosin, and scanned with an Aperio Scanscope (Aperio Technologies). Scanned sections were analyzed by a pathologist (A.M.) blinded to animal genotype.
Immunofluorescence analysis
Five μm sections of tissues as prepared above were stained with FITC-conjugated rabbit anti–human fibrin(ogen) antibodies (DAKO), polyclonal rabbit anti–human CD3 antibodies (DAKO), and polyclonal rabbit anti–human myeloperoxidase antibodies (DAKO; all cross-reactive with mouse antigens), as recommended by the manufacturers. CD3 and myeloperoxidase antibodies were visualized with Alexa Fluor 555–conjugated goat anti–rabbit IgG (Invitrogen). Imaging was performed with an LSM 700 Confocal Laser Scanning Microscope (Carl Zeiss).
Peritoneal inflammation model
Thioglycollate-induced peritonitis was performed with mice 8 to 10 weeks old as described.20 To quantitatively assess peritoneal neutrophil or macrophage accumulation, we humanely killed the mice at 4 or 72 hours, respectively, after injection of thioglycollate. Peritoneal lavage was collected, and the number of neutrophils or macrophages was quantified by flow cytometry analysis after staining for Gr-1 and CD11b using PE-Gr1– and APC-CD11b–conjugated antibodies (BD Biosciences).
Lung inflammation model
The lipopolysaccharide-induced lung inflammation assay was performed with mice 8 to 10 weeks old as described.21 Briefly, lipopolysaccharide serotype O55:B5 (Sigma-Aldrich; 2 μg/mouse) was administered intranasally. After an incubation period of 24 hours, the mice were humanely killed, and the bronchoalveolar lavage was collected using 1 mL of PBS, which was injected through a polyethylene tube placed into the upper airway after an incision into the trachea. We estimated the number of recruited neutrophils by counting the total cell number with a hemacytometer and by flow cytometry analysis using PE-Gr1– and APC-CD11b–conjugated antibodies.
Tissue repair assays
Cutaneous wound repair.
Full-thickness incisional skin wounds (15 mm) were generated in the interscapular dorsum. Healing was assessed by daily inspection of wounds by an investigator blinded to genotype of the mice, as described.22
Hepatic injury repair.
Assessment of hepatic repair was performed as described.23 Briefly, mice were injected intraperitoneally with 0.5 mL carbon tetrachloride (CCl4) per kilogram of body weight, as a 25% (vol/vol) solution in Guaranteed Value corn oil (Giant Food LLC), and humanely killed 2 and 8 days after injection. Liver sections were fixed for 16 to 18 hours in Z-fix, embedded into paraffin, sectioned, stained with hematoxylin & eosin, and scanned with an Aperio Scanscope (Aperio Technologies). Necrotic areas were quantified as a function of total liver area from these scanned files using Aperio ImageScope version 10.0.36.1805 software (Aperio Technologies) and the “Positive Pixel Count Algorithm V9” by an investigator who was blinded to animal genotype.
Results
Abrogation of uPA-uPAR interaction in mice
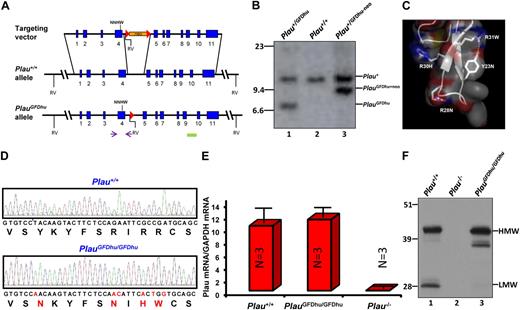
The uPA-uPAR interaction is primarily governed by the burial of the β-hairpin of the growth factor domain (GFD) of uPA into the central ligand-binding cavity of uPAR.16,24 In the present study, we exploited the well-known species selectivity of this interaction25 to generate a mouse strain (PlauGFDhu/GFDhu) in which the uPA-uPAR interaction was abrogated while maintaining the overall structure and catalytic function of uPA. This rational design was enabled by the recent disclosure of the structural basis underlying the species selectivity of this interaction.17 Specifically, we made 6 nucleotide changes in exon 4 of the Plau gene by homologous recombination in embryonic stem cells (Figure 1A-B), thereby introducing 4 amino acid substitutions (Y23N, R28N, R30H, and R31W) into the GFD of mouse uPA (Figure 1C). The 4 amino acid substitutions reduce the affinity of uPA for mouse uPAR by 400-fold17 while preserving the overall structure of the GFD.17 Furthermore, the substitutions that were chosen for the 4 amino acids endowed the quadruple-mutated mouse uPA with the ability to bind human uPAR with an affinity that is similar to that of human uPA (Kd, 0.33 vs 0.24nM,17 respectively), thereby allowing for selective reconstitution of the uPA-uPAR interaction by the introduction of human uPAR to PlauGFDhu/GFDhu cells, tissues or mice. Importantly, C57BL/6 ES cells were used for the generation of the novel knock-in mouse strain to prevent genetic background issues, such as cosegregation of linked genes, from confounding data interpretation in subsequent functional studies. Genomic analysis of PlauGFDhu/GFDhu mice confirmed the introduction of the 6-nucleotide substitution (Figure 1D). Quantitation of transcript levels showed that Plau gene transcription and pre-mRNA processing were not affected by the mutations in exon 4 or by the loxP site introduced into the intervening sequence between exons 4 and 5 by the targeting strategy (Figure 1E). In agreement with this, Western blot analysis of void urine (Figure 1F) showed that uPAY23N-R28N-R30H-R31W was expressed at normal levels. Interestingly, uPAY23N-R28N-R30H-R31W gave rise to the generation of less low-molecular-weight (LMW) uPA in void urine with a corresponding increase in a 38-kDa uPA species, revealing a different pattern of degradation. This altered pattern of uPA degradation was not a consequence of the loss of ability of uPAY23N-R28N-R30H-R31W to bind uPAR per se, as a 38-kDa uPA degradation product was not observed in the urine of uPAR-deficient mice.5,6 Importantly, the altered pattern of degradation did not affect the level of expression of uPAY23N-R28N-R30H-R31W (Figure 1F, compare lanes 1 and 3). Furthermore, the mutated uPA was functional in vivo, as it fully supported the 4 uPAR-independent functions of uPA that were analyzed in this study (maintenance of overall health, long-term survival, skin wound healing, and liver repair; see following sections).
Generation of the PlauGFDhu/GFDhu mouse strain. (A) Structure of targeting vector (top), wild-type Plau allele (middle), and targeted Plau allele with the neomycin cassette removed (bottom). Exons are indicated as blue boxes and intron sequences as black lines. Homologous recombination in ES cells introduced c.185T→A, c.201G→A, c.202A→C, c.207G→A, c.209C→T, and c.211A→G mutations into exon 4 and a neomycin selection cassette (orange) flanked by LoxP sites (red triangles) into intron 4. The neomycin cassette was removed by crossing germline chimeras to the Oz_Cre deletor strain (Ozgene Pty). The locations of the primers used for PCR screening of ES cell clones and genotyping of mice are indicated by arrows. NNHW indicates the position of the 4 codon changes in exon 4. Solid green box denotes location of Southern blot probe. Position of EcoRV restriction sites (RV) are indicated. (B) Southern blot of EcoRV digested tail DNA from Plau+/GFDhu (lane 1), Plau+/+ (lane 2), and Plau+/GFDhu-neo (lane 3) offspring of a PlauGFDhu/+ germline chimera crossed to an Oz_Cre deletor strain mouse. Positions of wildtype, targeted allele with the neomycin cassette, and targeted allele with the neomycin cassette removed are shown. Position of molecular weight markers (kb) are indicated at left. (C) Structure of the mouse uPA GFD. Combined surface (semitransparent), ribbon, and stick representation depicts the receptor-binding β-hairpin of the GFD of mouse uPA using the atomic coordinates 3LAQ17 and Pymol (DeLano Scientific). The 4 mutated amino acids (Y23N, R28N, R30H, R31W) are shown as sticks. (D) Sequence analysis of part of exon 4 (c.179-217) from Plau+/+ (top) and PlauGFDhu/GFDhu (bottom) mice confirms the introduction of the 6 nucleotide substitutions and 4 amino acid substitutions (red letters in nucleotide and amino acid sequence). (E) qPCR analysis of total RNA from Plau+/+ (left), PlauGFDhu/GFDhu (middle), and Plau−/− (right) mouse kidneys shows that Plau mRNA levels are not affected by the mutations in exon 4 or the LoxP site in intron 4. Data are shown as mean ± SEM, 3 mice per genotype. (F) Western blot of uPA in pooled samples (3-4 mice per group) of void urine from Plau+/+ (left), Plau−/− (middle), and PlauGFDhu/GFDhu (right) shows normal expression of high-molecular-weight (HMW, arrow) uPAY23N-R28N-R30H-R31W, reduced LWM (arrow) uPAY23N-R28N-R30H-R31W, and a corresponding increase in a higher-molecular-weight form processed in the linker region between the GFD and kringle domains. The position of molecular weight markers (kDa) is indicated at left.
Generation of the PlauGFDhu/GFDhu mouse strain. (A) Structure of targeting vector (top), wild-type Plau allele (middle), and targeted Plau allele with the neomycin cassette removed (bottom). Exons are indicated as blue boxes and intron sequences as black lines. Homologous recombination in ES cells introduced c.185T→A, c.201G→A, c.202A→C, c.207G→A, c.209C→T, and c.211A→G mutations into exon 4 and a neomycin selection cassette (orange) flanked by LoxP sites (red triangles) into intron 4. The neomycin cassette was removed by crossing germline chimeras to the Oz_Cre deletor strain (Ozgene Pty). The locations of the primers used for PCR screening of ES cell clones and genotyping of mice are indicated by arrows. NNHW indicates the position of the 4 codon changes in exon 4. Solid green box denotes location of Southern blot probe. Position of EcoRV restriction sites (RV) are indicated. (B) Southern blot of EcoRV digested tail DNA from Plau+/GFDhu (lane 1), Plau+/+ (lane 2), and Plau+/GFDhu-neo (lane 3) offspring of a PlauGFDhu/+ germline chimera crossed to an Oz_Cre deletor strain mouse. Positions of wildtype, targeted allele with the neomycin cassette, and targeted allele with the neomycin cassette removed are shown. Position of molecular weight markers (kb) are indicated at left. (C) Structure of the mouse uPA GFD. Combined surface (semitransparent), ribbon, and stick representation depicts the receptor-binding β-hairpin of the GFD of mouse uPA using the atomic coordinates 3LAQ17 and Pymol (DeLano Scientific). The 4 mutated amino acids (Y23N, R28N, R30H, R31W) are shown as sticks. (D) Sequence analysis of part of exon 4 (c.179-217) from Plau+/+ (top) and PlauGFDhu/GFDhu (bottom) mice confirms the introduction of the 6 nucleotide substitutions and 4 amino acid substitutions (red letters in nucleotide and amino acid sequence). (E) qPCR analysis of total RNA from Plau+/+ (left), PlauGFDhu/GFDhu (middle), and Plau−/− (right) mouse kidneys shows that Plau mRNA levels are not affected by the mutations in exon 4 or the LoxP site in intron 4. Data are shown as mean ± SEM, 3 mice per genotype. (F) Western blot of uPA in pooled samples (3-4 mice per group) of void urine from Plau+/+ (left), Plau−/− (middle), and PlauGFDhu/GFDhu (right) shows normal expression of high-molecular-weight (HMW, arrow) uPAY23N-R28N-R30H-R31W, reduced LWM (arrow) uPAY23N-R28N-R30H-R31W, and a corresponding increase in a higher-molecular-weight form processed in the linker region between the GFD and kringle domains. The position of molecular weight markers (kDa) is indicated at left.
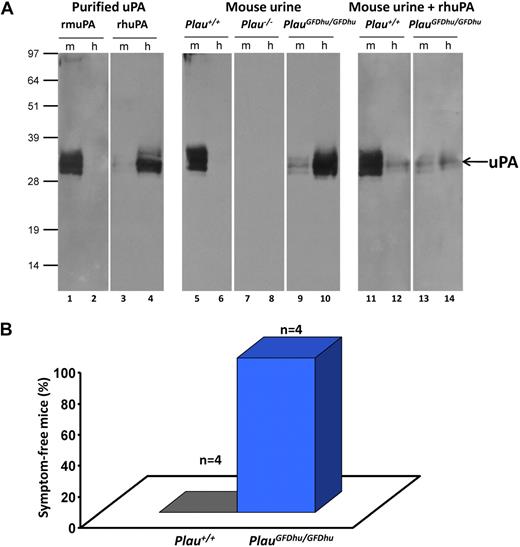
To demonstrate that uPAY23N-R28N-R30H-R31W produced by PlauGFDhu/GFDhu mice had an impaired ability to bind mouse uPAR and, correspondingly, had acquired the ability to bind human uPAR, a ligand blot was used (Figure 2A). Preparations of recombinant-soluble mouse or human uPAR were separated by SDS-PAGE, immobilized on PVDF membranes, and incubated with purified recombinant pro-uPAs or void urine from PlauGFDhu/GFDhu or Plau+/+ mice, followed by anti-uPA antibodies. As expected, void urine from Plau+/+ mice contained uPA that selectively bound mouse uPAR compared with human uPAR (Figure 2A lanes 5-6). Conversely, uPA from void urine of PlauGFDhu/GFDhu mice displayed negligible binding to mouse uPAR but bound to human uPAR, in agreement with a 400-fold reduction in affinity of the mutated uPA for mouse uPAR and an affinity for human uPAR that is similar to human uPA17 (Figure 2A lanes 9-10). This notion was further substantiated by the fact that receptor binding of murine uPA could only be competed by an excess human uPA added to the void urine from PlauGFDhu/GFDhu mice (Figure 2A lanes 11-14).
Specificity shift of uPAY23N-R28N-R30H-R31W from mouse to human uPAR. (A) The receptor-binding status of uPA in urine was tested by ligand blotting on PVDF membranes after SDS-PAGE of 0.5 μg purified mouse uPAR (m, lanes 1, 3, 5, 7, 9, 11, and 13) or human uPAR (h, lanes 2, 4, 6, 8, 10, 12, and 14). In lanes 1 to 4, the species selectivity of the uPAR-uPA interaction is demonstrated by incubating the blot with 1nM purified mouse pro-uPA (rmuPA, lanes 1-2) or human pro-uPA (rhuPA, lanes 3-4) spiked in urine from Plau−/− mice. In lanes 5 to 10, pools of void urine from Plau+/+ (n = 3; lanes 5-6), Plau−/− (n = 3; lanes 7-8), and PlauGFDhu/GFDhu (n = 7; lanes 9-10) mice were incubated with the uPAR blots. Bound uPA was detected with polyclonal rabbit anti-uPA antibodies, followed by swine anti–rabbit anti-IgG. In lanes 11-14, the specificities of these interactions were verified by incubating Plau+/+ (lanes 11-12) and PlauGFDhu/GFDhu (lanes 13-14) urine in the presence of 20nM recombinant human pro-uPA followed by a rabbit polyclonal antibody specific for mouse uPA. All urine samples were diluted 20-fold in PBS containing 0.05% (vol/vol) Tween 20 before incubation overnight at 4°C. The position of molecular weight markers (kDa) is indicated at left. (B) 4 adult Plau+/+ (left column) and 4 PlauGFDhu/GFDhu (right column) mice were injected intraperitoneally with 8 mg/kg PrAg-U2 and 10 mg/kg FP59. The health of the mice was determined by an investigator unaware of animal genotype. The mice were then humanely killed when moribund (P = .014; Fisher exact test).
Specificity shift of uPAY23N-R28N-R30H-R31W from mouse to human uPAR. (A) The receptor-binding status of uPA in urine was tested by ligand blotting on PVDF membranes after SDS-PAGE of 0.5 μg purified mouse uPAR (m, lanes 1, 3, 5, 7, 9, 11, and 13) or human uPAR (h, lanes 2, 4, 6, 8, 10, 12, and 14). In lanes 1 to 4, the species selectivity of the uPAR-uPA interaction is demonstrated by incubating the blot with 1nM purified mouse pro-uPA (rmuPA, lanes 1-2) or human pro-uPA (rhuPA, lanes 3-4) spiked in urine from Plau−/− mice. In lanes 5 to 10, pools of void urine from Plau+/+ (n = 3; lanes 5-6), Plau−/− (n = 3; lanes 7-8), and PlauGFDhu/GFDhu (n = 7; lanes 9-10) mice were incubated with the uPAR blots. Bound uPA was detected with polyclonal rabbit anti-uPA antibodies, followed by swine anti–rabbit anti-IgG. In lanes 11-14, the specificities of these interactions were verified by incubating Plau+/+ (lanes 11-12) and PlauGFDhu/GFDhu (lanes 13-14) urine in the presence of 20nM recombinant human pro-uPA followed by a rabbit polyclonal antibody specific for mouse uPA. All urine samples were diluted 20-fold in PBS containing 0.05% (vol/vol) Tween 20 before incubation overnight at 4°C. The position of molecular weight markers (kDa) is indicated at left. (B) 4 adult Plau+/+ (left column) and 4 PlauGFDhu/GFDhu (right column) mice were injected intraperitoneally with 8 mg/kg PrAg-U2 and 10 mg/kg FP59. The health of the mice was determined by an investigator unaware of animal genotype. The mice were then humanely killed when moribund (P = .014; Fisher exact test).
We next verified that uPAY23N-R28N-R30H-R31W had lost functional binding to uPAR in vivo (Figure 2B). We have previously generated a modified anthrax toxin that requires proteolytic activation by uPA on the cell surface for cytotoxicity and showed that activation of the mutated toxin in vitro and in vivo is dependent on the expression of uPA and uPAR.18,26 As reported previously, Plau+/+ mice became terminally ill within 48 hours after treatment with 8 mg/kg of this toxin (5 times the maximal tolerated dose in C57BL/6J mice).18 However, as previously described for Plau−/− and Plaur−/− mice,18 littermate PlauGFDhu/GFDhu mice treated in parallel with the toxin remained entirely unaffected, showing that the uPA-uPAR interaction is functionally abrogated in vivo in mice expressing uPAY23N-R28N-R30H-R31W.
Chronic inflammation caused by loss of uPA-uPAR interaction
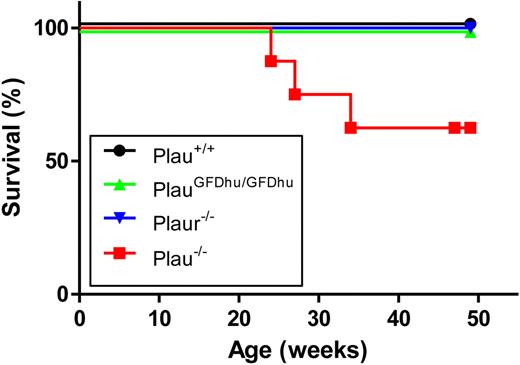
To assess the long-term consequences of the specific abrogation of uPA binding to uPAR, we next set up prospective cohorts of PlauGFDhu/GFDhu and littermate Plau+/+ mice and monitored their outward appearance and survival for 1 year. Cohorts of isogenic Plau−/− and Plaur−/− mice were also included for comparison. The 1-year survival of PlauGFDhu/GFDhu mice was similar to that of littermate Plau+/+ mice (Figure 3). In contrast, only 63% of the Plau−/− mice (5/8) survived for 1 year, and these surviving mice were emaciated when they were humanely killed (body weight, 69%-86% of Plau+/+), thus underscoring the essential role of uPA in maintaining long-term mouse health independent of its binding to uPAR. Plasminogen-deficient mice display a similar wasting syndrome that is caused by impaired fibrinolysis and first becomes apparent at ages 2 to 3 months.27
Survival of mice with abrogated uPA-uPAR interaction. The 1-year survival of a prospective cohort of Plau+/+ (n = 8), PlauGFDhu/GFDhu (n = 9), Plaur−/− (n = 7), and Plau−/− (n = 7) mice. P < .01, Plau−/− versus other genotypes, log rank test, 2-tailed.
Survival of mice with abrogated uPA-uPAR interaction. The 1-year survival of a prospective cohort of Plau+/+ (n = 8), PlauGFDhu/GFDhu (n = 9), Plaur−/− (n = 7), and Plau−/− (n = 7) mice. P < .01, Plau−/− versus other genotypes, log rank test, 2-tailed.
We next subjected the PlauGFDhu/GFDhu and littermate Plau+/+ mice to a detailed histopathologic examination by an investigator who was blinded to animal genotype. Interestingly, mice with abrogated uPA-uPAR interaction could be readily distinguished from their Plau+/+ littermates by the abundance of leukocytic infiltrates in their livers (Figure 4A-B). Indeed, a quantitative histomorphometric analysis showed that inflammatory foci in the liver were 4-fold more frequent in PlauGFDhu/GFDhu mice than in their littermate Plau+/+ mice (Figure 4B).
Chronic inflammation caused by abrogation of uPA-uPAR interaction. (A) Histologic appearance of the livers of 1-year-old Plau+/+ (top left panel), PlauGFDhu/GFDhu (top right panel), Plaur−/− (bottom left panel), and Plau−/− (bottom right panel) mice. Focal leukocyte infiltrates (examples with arrows) are increased in mice with abrogated uPA-uPAR interaction, loss of uPAR, or loss of uPA. Size bars, 50 μm. (B) Enumeration of inflammatory foci in the livers of Plau+/+ (left), PlauGFDhu/GFDhu (second left), Plaur−/− (second right), and Plau−/− (right) mice. Symbols represent individual mice. Horizontal bars show average values. P values were determined by the Mann-Whitney U test, 2-tailed. (C) Examples of immunostaining for neutrophils (myeloperoxidase, top panels) and T lymphocytes (CD3, bottom panels) within inflammatory foci from Plau+/+ (left panels), PlauGFDhu/GFDhu (second panels from left), Plaur−/− (second panels from right), and Plau−/− (right panels) showing prominent infiltration of neutrophils and T lymphocytes into tissues from mice with abrogated uPA-uPAR interaction, but not into mice with unchallenged uPA-uPAR interaction. Size bars, 20 μm.
Chronic inflammation caused by abrogation of uPA-uPAR interaction. (A) Histologic appearance of the livers of 1-year-old Plau+/+ (top left panel), PlauGFDhu/GFDhu (top right panel), Plaur−/− (bottom left panel), and Plau−/− (bottom right panel) mice. Focal leukocyte infiltrates (examples with arrows) are increased in mice with abrogated uPA-uPAR interaction, loss of uPAR, or loss of uPA. Size bars, 50 μm. (B) Enumeration of inflammatory foci in the livers of Plau+/+ (left), PlauGFDhu/GFDhu (second left), Plaur−/− (second right), and Plau−/− (right) mice. Symbols represent individual mice. Horizontal bars show average values. P values were determined by the Mann-Whitney U test, 2-tailed. (C) Examples of immunostaining for neutrophils (myeloperoxidase, top panels) and T lymphocytes (CD3, bottom panels) within inflammatory foci from Plau+/+ (left panels), PlauGFDhu/GFDhu (second panels from left), Plaur−/− (second panels from right), and Plau−/− (right panels) showing prominent infiltration of neutrophils and T lymphocytes into tissues from mice with abrogated uPA-uPAR interaction, but not into mice with unchallenged uPA-uPAR interaction. Size bars, 20 μm.
To explore the nature of these leukocytic infiltrates, we next performed immunostaining using markers of acute inflammation (myeloperoxidase, labeling neutrophils) and chronic inflammation (CD3, labeling T-lymphocytes). This analysis demonstrated a prominent contribution of lymphocytes to the leukocytic infiltrates and a minor contribution from neutrophils, thus revealing a novel role of the uPA-uPAR interaction in suppressing chronic tissue inflammation (Figure 4C). Analysis of Plaur−/− mice (Figure 4A) revealed inflammatory foci in the liver that were similar in number to those of PlauGFDhu/GFDhu mice (Figure 4B) and also were chronic in nature, as determined by the prominence of lymphocytes within the lesions (Figure 4C). Plau−/− mice displayed similar foci of chronic inflammation, which were 2.5-fold more abundant than foci in PlauGFDhu/GFDhu or Plaur−/− mice, although this difference did not reach statistical significance. Notably, the small and infrequent inflammatory foci found in Plau+/+ mice differed in cellular composition from foci in PlauGFDhu/GFDhu, Plaur−/−, and Plau−/− mice by having neutrophils as the primary cellular component (Figure 4C). All Plau−/− mice displayed extensive inflammation of the trachea with basement membrane hyperplasia and desquamation of the tracheal epithelium, whereas mild to moderate inflammation of the trachea was found in 2 (22%) of 9 of PlauGFDhu/GFDhu, 3 (43%) of 7 Plaur−/−, and 0 (0%) of 8 of Plau+/+ mice (data not shown). Other organs of 1-year old PlauGFDhu/GFDhu, Plaur−/−, and Plau−/− mice did not display histopathologic features that were different from that observed in cohabitating Plau+/+ mice, with the exception of reduced body fat stores and hepatic glycogen in Plau−/− mice (data not shown).
Leukocyte accumulation is unaffected by loss of uPA binding to uPAR in standard models of inflammation
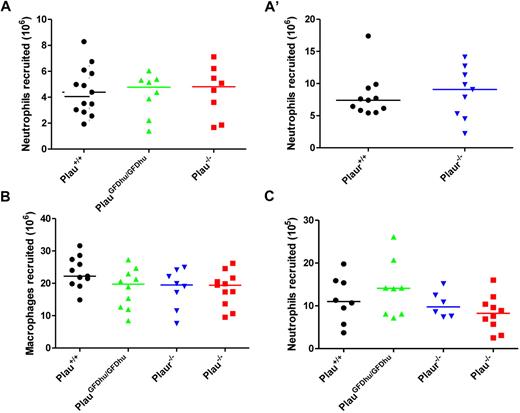
To identify the intrinsic defects that caused chronic inflammation in mice with abrogated uPA binding to uPAR, we first determined the specific contribution of the uPA-uPAR interaction to the regulation of leukocyte accumulation at sites of inflammation using 2 well-established models. We first injected thioglycollate into the peritoneum of PlauGFDhu/GFDhu and Plau+/+ littermate mice and enumerated the neutrophils and macrophages by peritoneal lavage after 4 and 72 hours, respectively. Both neutrophil (Figure 5A and A′) and macrophage (Figure 5B) recruitment were unaffected by the absence of uPA binding to uPAR, as shown by the appearance of similar numbers of the 2 leukocyte populations in PlauGFDhu/GFDhu and Plau+/+mice. Congruent with these findings, the complete ablation of either uPA or uPAR also did not affect either neutrophil or macrophage recruitment to the peritoneum in response to thioglycollate (Figure 5A, 5A′, and 5B). To establish if these findings could be replicated in a different anatomic context and using a different inflammatory stimulus, we next determined the effect of the loss of uPA binding to uPAR on neutrophil accumulation to the lungs by intratracheal installation of lipopolysaccharide to PlauGFDhu/GFDhu and littermate Plau+/+ mice followed by enumeration of the elicited neutrophils by bronchoalveolar lavage. Again, the abrogation of the uPA-uPAR interaction did not affect inflammatory cell accumulation, and neither did the complete loss of either uPA or uPAR (Figure 5C).
Loss of uPA-uPAR interaction does not affect leukocyte accumulation in standard models of inflammation. (A and A') Peritoneal neutrophils (CD11b−Gr1+) at 4 hours and (B) macrophages (CD11b+Gr1−) at 72 hours after thioglycollate injection of Plau+/+ (left), PlauGFDhu/GFDhu (second left), Plaur−/− (third left), and Plau−/− (right) mice. Experiments in A and A' were performed on separate days. (C) Lipopolysaccharide-elicited lung neutrophils (CD11b−Gr1+) at 24 hours in Plau+/+ (left), PlauGFDhu/GFDhu (second left), Plaur−/− (third left), and Plau−/− (right) mice. Symbols represent individual mice. Horizontal bars show average values. P = N.S., all genotypes versus each other, Mann-Whitney U test, 2-tailed.
Loss of uPA-uPAR interaction does not affect leukocyte accumulation in standard models of inflammation. (A and A') Peritoneal neutrophils (CD11b−Gr1+) at 4 hours and (B) macrophages (CD11b+Gr1−) at 72 hours after thioglycollate injection of Plau+/+ (left), PlauGFDhu/GFDhu (second left), Plaur−/− (third left), and Plau−/− (right) mice. Experiments in A and A' were performed on separate days. (C) Lipopolysaccharide-elicited lung neutrophils (CD11b−Gr1+) at 24 hours in Plau+/+ (left), PlauGFDhu/GFDhu (second left), Plaur−/− (third left), and Plau−/− (right) mice. Symbols represent individual mice. Horizontal bars show average values. P = N.S., all genotypes versus each other, Mann-Whitney U test, 2-tailed.
uPA supports tissue repair independent of binding to uPAR
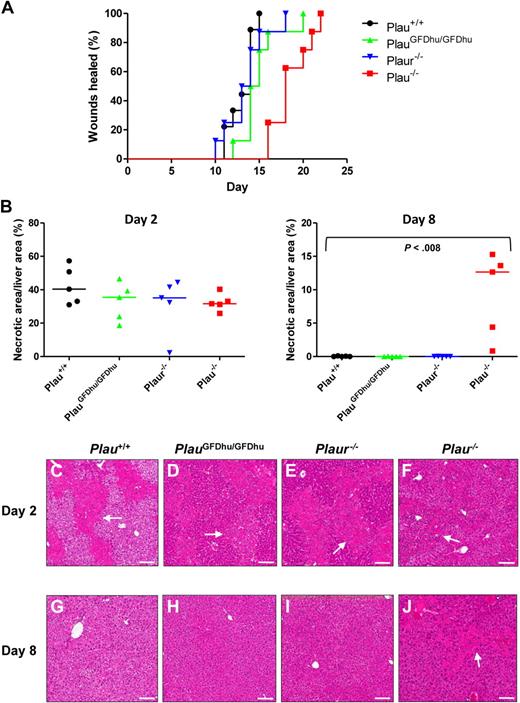
To determine whether the systemic inflammation that befalls PlauGFDhu/GFDhu mice is secondary to a general defect in the healing of injured tissues, incisional skin wounds were generated in PlauGFDhu/GFDhu and littermate Plau+/+ mice, and the time required to complete healing was determined (Figure 6A). Abrogation of the uPA-uPAR interaction did not delay skin wound healing (healing times: PlauGFDhu/GFDhu, 15 ± 0.8 days; Plau+/+ mice, 13 ± 0.5 days, P = N.S.). Skin wound healing was also unaffected by the complete loss of uPAR (healing times: Plaur−/−, 14 ± 0.9 days). In contrast, wholesale loss of uPA significantly impaired skin wound healing (healing time: Plau−/−, 19 ± 0.8, P < .001 vs Plau+/+), showing that uPA promotes wound repair independent of binding to its receptor. To elaborate this finding using a different injury type and anatomic context, we next administered carbon tetrachloride intraperitoneally to mice of each genotype to induce hepatic necrosis.23 The extent of liver injury induced by this treatment did not differ between mice of the 4 genotypes when assessed at day 2 (Figure 6B left panel, C-F). However, whereas the necrotic debris was cleared from the livers of Plau+/+ mice, PlauGFDhu/GFDhu, and Plaur−/− mice at day 8, substantial areas of necrosis persisted in Plau−/− mouse livers (Figure 6B right panel, G-J). Taken together, these studies show that uPA promotes tissue repair independent of binding to its cellular receptor.
Loss of uPA binding to uPAR does not affect tissue repair. (A) Rate of healing of 1.5-cm incisional skin wounds in Plau+/+ (black, N = 10), PlauGFDhu/GFDhu (green N = 10), Plaur−/− (blue, N = 10), and Plau−/− (red, N = 9) mice. Loss of uPA, but not uPA binding, to uPAR or loss of uPAR delays wound healing. P < .001 Plau−/− versus Plau+/+, PlauGFDhu/GFDhu or Plaur−/−, log-rank test, 2-tailed. (B) Area of hepatic necrosis 2 (left panel) and 8 (right panel) days after carbon tetrachloride–induced liver injury of Plau+/+ (black circles), PlauGFDhu/GFDhu (green triangles), Plaur−/− (blue triangles), and Plau−/− (red squares). A total of 5 mice were analyzed per genotype and time point. The extent of hepatic injury is similar in mice of all genotypes at day 2. At day 8, necrotic areas are completely cleared in Plau+/+, PlauGFDhu/GFDhu, and Plaur−/− livers, but not in Plau−/− livers. The P value was determined by the Mann-Whitney U test, 2-tailed. (C-J) Representative examples of the histologic appearance of livers from Plau+/+ (C,G), PlauGFDhu/GFDhu (D,H), Plaur−/− (E,I), and Plau−/− (F,J) mice 2 (C-F) and 8 (G-J) days after carbon tetrachloride administration. White arrows show examples of necrosis. Size bars, 100 μm.
Loss of uPA binding to uPAR does not affect tissue repair. (A) Rate of healing of 1.5-cm incisional skin wounds in Plau+/+ (black, N = 10), PlauGFDhu/GFDhu (green N = 10), Plaur−/− (blue, N = 10), and Plau−/− (red, N = 9) mice. Loss of uPA, but not uPA binding, to uPAR or loss of uPAR delays wound healing. P < .001 Plau−/− versus Plau+/+, PlauGFDhu/GFDhu or Plaur−/−, log-rank test, 2-tailed. (B) Area of hepatic necrosis 2 (left panel) and 8 (right panel) days after carbon tetrachloride–induced liver injury of Plau+/+ (black circles), PlauGFDhu/GFDhu (green triangles), Plaur−/− (blue triangles), and Plau−/− (red squares). A total of 5 mice were analyzed per genotype and time point. The extent of hepatic injury is similar in mice of all genotypes at day 2. At day 8, necrotic areas are completely cleared in Plau+/+, PlauGFDhu/GFDhu, and Plaur−/− livers, but not in Plau−/− livers. The P value was determined by the Mann-Whitney U test, 2-tailed. (C-J) Representative examples of the histologic appearance of livers from Plau+/+ (C,G), PlauGFDhu/GFDhu (D,H), Plaur−/− (E,I), and Plau−/− (F,J) mice 2 (C-F) and 8 (G-J) days after carbon tetrachloride administration. White arrows show examples of necrosis. Size bars, 100 μm.
uPA binding to uPAR promotes fibrin surveillance
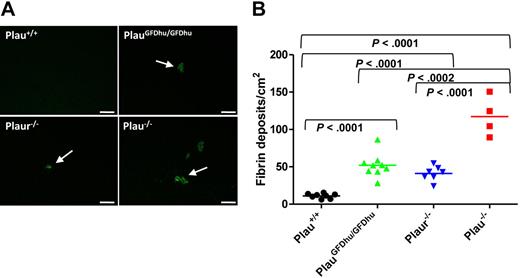
Plasmin is the major fibrinolytic protease in vivo, and uPA is one of 2 principal plasminogen activators engaged in fibrinolysis.4,28,–30 Therefore, we next assessed the specific contribution of uPA binding to uPAR to fibrin surveillance by immunofluorescence using fibrin-specific antibodies. Interestingly, the abrogation of uPA binding to uPAR compromised the capacity to prevent fibrin accumulation, as assessed by the large increase in fibrin foci in hepatic tissues from 1-year-old PlauGFDhu/GFDhu mice compared with littermate Plau+/+ mice (Figure 7A-B). This result was unexpected, as fibrin deposition has not been reported in uPAR-deficient mice in the absence of additional fibrinolytic deficits.5,6,9 However, the 1-year-old Plaur−/− mice enrolled in the prospective cohort that was examined in the current study also showed liver fibrin deposits that were qualitatively (Figure 7A) and quantitatively (Figure 7B) identical to those observed in PlauGFDhu/GFDhu mice, thus supporting a role of uPA binding to uPAR in fibrin surveillance. Plau−/− mice displayed similar fibrin deposits that were 2.5-fold more frequent in number than fibrin deposits in PlauGFDhu/GFDhu and Plaur−/− mice (Figure 7A-B). Fibrin deposits were also observed in the trachea of Plau−/− mice, which were not detectable in either PlauGFDhu/GFDhu or Plaur−/− mice (data not shown).
Fibrin accumulation caused by loss of uPA-uPAR interaction. (A) Representative low-magnification images of liver sections from Plau+/+ (top left), PlauGFDhu/GFDhu (top right), Plaur−/− (bottom left), and Plau−/− (bottom right) mice stained with fibrin-specific antibodies showing fibrin deposits. Size bars, 100 μm. (B) Enumeration of fibrin foci in the livers of Plau+/+ (left), PlauGFDhu/GFDhu (second left), Plaur−/− (second right), and Plau−/− (right) mice. Symbols represent individual mice. Horizontal bars show average values. P values were determined by the Student t test, 2-tailed.
Fibrin accumulation caused by loss of uPA-uPAR interaction. (A) Representative low-magnification images of liver sections from Plau+/+ (top left), PlauGFDhu/GFDhu (top right), Plaur−/− (bottom left), and Plau−/− (bottom right) mice stained with fibrin-specific antibodies showing fibrin deposits. Size bars, 100 μm. (B) Enumeration of fibrin foci in the livers of Plau+/+ (left), PlauGFDhu/GFDhu (second left), Plaur−/− (second right), and Plau−/− (right) mice. Symbols represent individual mice. Horizontal bars show average values. P values were determined by the Student t test, 2-tailed.
Chronic inflammation caused by loss of uPA-uPAR interaction is secondary to fibrin deposition
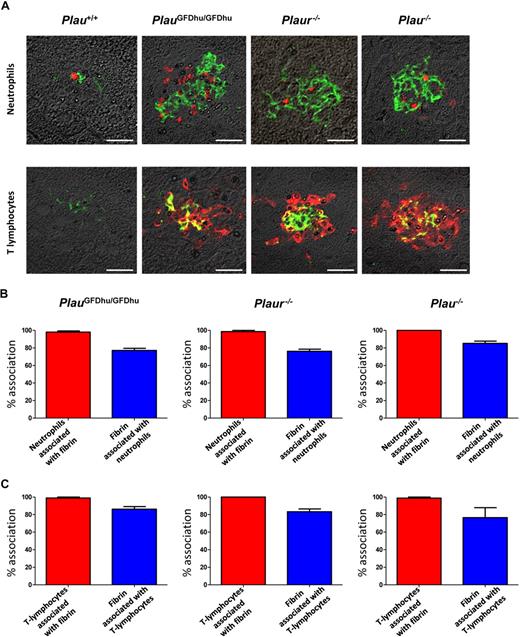
Fibrin is endowed with potent proinflammatory properties and its persistence in tissues can lead to chronic inflammation (see “Discussion”). This property of fibrin, when combined with the unaltered leukocyte accumulation in response to inflammatory stimuli and the uncompromised tissue repair observed in PlauGFDhu/GFDhu mice, suggested that the systemic inflammation caused by abrogation of the uPA-uPAR interaction in these mice could be secondary to a compromised ability to eliminate fibrin deposits in tissues. To investigate this, we combined fibrin immunofluorescence with staining for neutrophils (myeloperoxidase; Figure 8A) and lymphocytes (CD3; Figure 8A) and performed quantitative studies of the association between fibrin deposition and leukocytic tissue infiltrates (Figure 8B-C). Interestingly, this survey revealed a near-complete colocalization of neutrophils and T lymphocytes with fibrin deposits in PlauGFDhu/GFDhu, Plaur−/−, and Plau−/− mice (Figure 8B-C). Accordingly, the majority of fibrin deposits in PlauGFDhu/GFDhu, Plaur−/−, and Plau−/− mice was associated with neutrophils and T lymphocytes (Figure 8B-C).
Inflammation caused by uPA-uPAR abrogation is secondary to fibrin deposition. (A) Examples of immunofluorescence using fibrin antibodies (green) combined with antibodies against neutrophils (myeloperoxidase, top panels) and T lymphocytes (CD3, bottom panels; red) of the livers from 1-year-old Plau+/+ (left panels), PlauGFDhu/GFDhu (2 left panels), Plaur−/− (2 right panels), and Plau−/− (right panels). Size bars, 20 μm. (B) Enumeration of the percentage of neutrophils associated with fibrin (red bars) and the percentage of fibrin deposits associated with neutrophils (blue bars) in the livers from PlauGFDhu/GFDhu (left), Plaur−/− (middle), and Plau−/− (right) panels. (C) Enumeration of the percentage of T lymphocytes associated with fibrin (red bars) and the percentage of fibrin deposits associated with T lymphocytes (blue bars) in the livers from PlauGFDhu/GFDhu (left), Plaur−/− (middle), and Plau−/− (right) panels.
Inflammation caused by uPA-uPAR abrogation is secondary to fibrin deposition. (A) Examples of immunofluorescence using fibrin antibodies (green) combined with antibodies against neutrophils (myeloperoxidase, top panels) and T lymphocytes (CD3, bottom panels; red) of the livers from 1-year-old Plau+/+ (left panels), PlauGFDhu/GFDhu (2 left panels), Plaur−/− (2 right panels), and Plau−/− (right panels). Size bars, 20 μm. (B) Enumeration of the percentage of neutrophils associated with fibrin (red bars) and the percentage of fibrin deposits associated with neutrophils (blue bars) in the livers from PlauGFDhu/GFDhu (left), Plaur−/− (middle), and Plau−/− (right) panels. (C) Enumeration of the percentage of T lymphocytes associated with fibrin (red bars) and the percentage of fibrin deposits associated with T lymphocytes (blue bars) in the livers from PlauGFDhu/GFDhu (left), Plaur−/− (middle), and Plau−/− (right) panels.
These findings provide strong evidence that the spontaneous accumulation of inflammatory cells in tissues associated with the selective loss of the uPA-uPAR interaction was secondary to impaired fibrin surveillance. To investigate this phenomenon in greater detail, we next performed 3-dimensional reconstructions of foci of inflammation after the visualization of T-lymphocytes and fibrin by immunofluorescence (supplemental Video 1). Analysis of multiple inflammatory foci from PlauGFDhu/GFDhu, Plaur−/−, and Plau−/− mice showed that the lymphocytic infiltrates uniformly were organized around a central core of fibrin, thus further supporting that the chronic inflammation caused by the abrogation of the uPA-uPAR interaction is secondary to an intrinsic defect in fibrin homeostasis.
Discussion
Fibrinogen is a circulating hemostatic factor whose conversion into fibrin is essential for stemming bleeding after vessel rupture, as well as for controlling the initial stages of bacterial infection. In its latter capacity, fibrin serves as a nondiffusible scaffold that attracts and activates leukocytes through the ligation of fibrin(ogen)-binding integrin receptors.31,–33 However, the capacity to dissolve deposited fibrin in a timely manner is equally essential to maintaining health as the capacity to convert fibrinogen to fibrin. This is well illustrated by the progressive inflammation-associated, fibrin-dependent multiorgan pathology and impaired tissue regenerative capacity that is observed in humans and mice deficient in the key fibrinolytic protease zymogen plasminogen27,29,30,34,,,,,,,–42 and by studies in humans and animals showing that extravascular fibrin deposition exacerbates the morbidity of a wide range of chronic human diseases, including tissue fibrosis, muscular dystrophy, rheumatoid arthritis, and multiple sclerosis, in large part through its ability to cause persistent inflammation.43,,,,,–49
Previous studies have suggested important roles of uPAR in cell adhesion, cell migration, cell proliferation, cell differentiation, and cell survival (Smith and Marshall2 ; Blasi and Carmeliet3 ; supplemental References 1-22). In this paper, we identified yet another function for uPAR in suppression of fibrin-associated chronic inflammation. Thus, our novel mouse strain, in which the uPA-uPAR interaction is selectively abrogated by subtle point mutations in the uPA GFD, developed a syndrome that was characterized by fibrin deposits that were infiltrated by inflammatory cells of both myeloid and lymphoid origin. This syndrome was mirrored in isogenic mice deficient in uPAR and was qualitatively similar, but quantitatively exacerbated, in isogenic uPA–deficient mice. Loss of the uPA-uPAR interaction was not associated with intrinsic changes in the kinetics of leukocyte accumulation in response to inflammatory stimuli or was a consequence of defective tissue repair, as determined in standard models of inflammation, and wound healing. As uPA is one of the 2 principal activators of plasminogen, and uPAR promotes uPA-mediated plasminogen activation on the cell surface,50 the most straightforward interpretation of the data from the current study, therefore, is that loss of uPA binding to uPAR compromises a uPA-dependent cell surface plasminogen activation pathway that is engaged in fibrin surveillance. This pathway cannot be fully compensated for by either uPAR-independent plasminogen activation by uPA9 or by activation of plasminogen by tPA.4 Thus, our study identifies a principal in vivo role of the uPA-uPAR interaction in cell-associated fibrin surveillance.
A prior analysis of mice deficient in uPAR conducted by 2 independent groups failed to detect excessive fibrin deposition, leading to the conclusion that fibrin surveillance by uPA is predominantly a uPAR-independent process.5,6 The most likely explanation for the divergent findings is that only younger mice (age 4.5 weeks5 and ages 18-29 weeks6 ) were examined in the 2 prior studies, whereas the mice in the current study were analyzed at age 1 year. Differences in genetic background and animal housing conditions are other factors potentially contributing to discrepancies between the current study and the 2 previous studies.
During the past 2 decades, uPAR has emerged as a potential orchestrator of multiple cellular events in the context of numerous physiologic and pathologic processes.2,3 In addition to our identification of a principal in vivo role of the uPA-uPAR interaction in suppression of fibrin-associated inflammation, the novel mouse strain developed in this study will be a most valuable tool for further functional dissection of this remarkable cell-surface molecule.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Drs Silvio Gutkind and Mary Jo Danton for critically reviewing this manuscript.
This study was supported by the National Institute of Dental and Craniofacial Research, National Institutes of Health, and National Institute of Cancer Intramural Research Programs, and grants from the Danish National Research Foundation (Danish-Chinese Center for Proteases and Cancer), and the Lundbeck Foundation. PlauGFDhu/GFDhu mice can be obtained by contacting Thomas Bugge (thomas.bugge@nih.gov).
National Institutes of Health
Authorship
Contribution: B.M.C., E.Y.C., H.G., A.L.B., B.M.C., T.C., S.L., A.M., M.P., and S.H.L. designed and performed research and analyzed data; and T.H.B. designed and performed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation of T.C. is Division of Vascular Inflammation, Diabetes and Kidney, Department of Medicine III and Institute of Physiology, Dresden University of Technology, Germany.
Correspondence: Thomas H. Bugge, Proteases and Tissue Remodeling Section, Oral and Pharyngeal Cancer Branch, National Institute of Dental and Craniofacial Research, National Institutes of Health, 30 Convent Dr, Rm 211, Bethesda, MD 20892; e-mail: thomas.bugge@nih.gov.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal