In this issue of Blood, Ballabio and colleagues performed microRNA (miRNA) profiling in Sezary syndrome (SzS) and identified that miR-233 can distinguish T cells from patients with SzS from healthy controls and from patients with mycosis fungoides.1 In addition, they identified that down-regulation of miR-342 may play a role in the pathogenesis of SzS through inhibition of apoptosis. The investigators also identified a novel mechanism of regulation of miR-342 via binding of miR-199a* to the host gene of miR-342, EVL.
SzS is the leukemic variant of the most common subtype of cutaneous T-cell lymphoma (CTCL), mycosis fungoides, and constitutes 2.5% of CTCL. It is characterized by generalized erythroderma, lymphadenopathy, and presence of 5% or more of CD4+ malignant T cells with cerebriform nuclei (Sezary cells). Although common genetic aberrations and defects in cytokine response and immune regulation have been identified, the molecular biology of the disease has not been well established.2
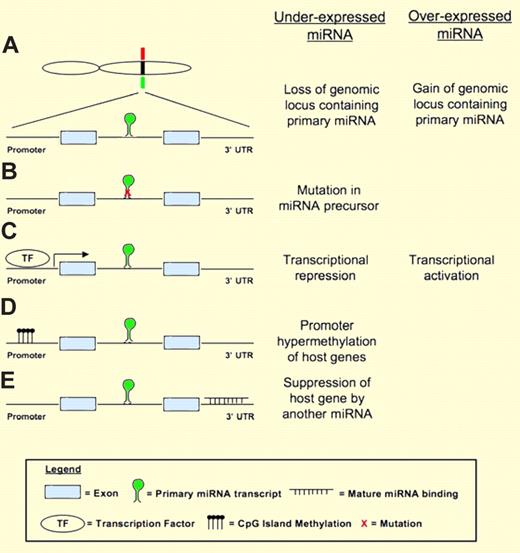
Mechanisms of miRNA dysregulation in cancer. The example shown here to illustrate the different mechanisms has the miRNA located in the intron of a host gene containing 2 exons. (A) A miRNA may be underexpressed or overexpressed due to deletion (green bar) or gain (red bar) of the genomic locus containing the miRNA. (B) Mutation in the miRNA precursor that affects the processing of miRNA may lead to underexpression of the miRNA. (C) Transcriptional repression (eg, by MYC) or activation (eg, by p53) by transcription factors may lead to underexpression or overexpression of miRNA. (D) miRNA expression may be suppressed by promoter methylation. (E) A novel mechanism identified by Ballabio and colleagues where miRNA expression is suppressed by other miRNA inhibiting the expression of the host gene.
Mechanisms of miRNA dysregulation in cancer. The example shown here to illustrate the different mechanisms has the miRNA located in the intron of a host gene containing 2 exons. (A) A miRNA may be underexpressed or overexpressed due to deletion (green bar) or gain (red bar) of the genomic locus containing the miRNA. (B) Mutation in the miRNA precursor that affects the processing of miRNA may lead to underexpression of the miRNA. (C) Transcriptional repression (eg, by MYC) or activation (eg, by p53) by transcription factors may lead to underexpression or overexpression of miRNA. (D) miRNA expression may be suppressed by promoter methylation. (E) A novel mechanism identified by Ballabio and colleagues where miRNA expression is suppressed by other miRNA inhibiting the expression of the host gene.
miRNA are small noncoding RNAs of approximately 22 nucleotides that target mRNAs and function as posttranslational regulators. Generally, miRNA binds to the 3′ untranslated region of target mRNA via complementary binding. Binding typically leads to either translational repression or mRNA degradation. miRNA can be located on introns or exons of the coding (host gene) or noncoding transcriptional unit.3
Aberrant expression of miRNA has been described in different cancers.4 Global reduction in miRNA biogenesis is tumorigenic,5 and may be due to deletion of DICER1, a key protein involved in miRNA biogenesis and a haploinsufficient tumor suppressor gene.6 miRNAs are also located in genomic loci commonly aberrant in cancers and may be deregulated as a result of gain or loss of genomic loci.7 One miRNA may affect multiple targets and have wide ranging impact on gene and protein expression. Although their impact on an individual target may be subtle, the combined effect on a synergistic network of genes may have significant consequence. They have been shown to be responsible for mediating some of the downstream effect of important oncogenes and tumor suppressor genes such and MYC and p53. Overall, the body of evidence suggests miRNA are important to tumor biology.
Ballabio and colleagues identify a single miRNA, miR-223, that could distinguish SzS from healthy controls and mycosis fungoides with 90% accuracy, suggesting that this may be a useful diagnostic marker. While exciting, how this performs in relation to previously defined gene expression signatures will need to be formally addressed. Whether miR-223 can be used to identify cases currently difficult to diagnose will be a critical test of its clinical utility. The process by which this diagnostic biomarker was identified highlights the difficulty in validating predictive signatures. The attrition from the initial 10 discriminating miRNA to just one suggests there are inconsistencies across different platforms (microarray vs reverse transcriptase–polymerase chain reaction) and datasets. It would be important to independently validate this result to determine the wider applicability of this result.
How are miRNA dysregulated in SzS? Ballabio et al extrapolated data from their previous studies using different datasets showing that many of the dysregulated miRNAs are located in loci of gains and losses previously identified; in addition, many of the predicted targets have abnormal gene expression suggesting miRNA dysregulation in SzS has functional consequences. Although compelling, the DNA copy-miRNA-mRNA correlation should be performed on the same samples before one can confirm a cause-and-effect relationship.
In my opinion, the most important contribution of the current study is the identification of a novel mechanism by which miRNA expression is regulated. Various mechanisms that may lead to abnormal miRNA expression in cancer have been identified4 (Figure 1). The current finding that miRNA expression can be regulated by other miRNAs through modulation of host gene expression expands the repertoire. This was robustly verified experimentally for miR-342, which was also shown to have tumor suppressor properties in SzS. Future studies should be directed at identifying addition regulator miRNAs. This will help define the true prevalence of this novel mechanism of miRNA regulation.
The relationship between miRNAs and genes is becoming complex, but it is important to rationally dissect this maze. miRNA may affect gene expression, yet certain genes—for example, transcription factors and genes involved in DNA methylation who are themselves targets of miRNA—may then affect miRNA expression. Analogous to identifying the driving mutations from the passenger mutations in the large number of somatic mutations identified in sequencing studies in cancer, we need a comprehensive view of what constitutes primary events and secondary events in dysregulated miRNA expression in cancer. This will become increasingly important in the future as miRNA-based gene therapy becomes reality. We will need to identify the critical nodes in this complex network for therapeutic targeting. To this end, the work of Ballabio and colleagues will have wider implication in our understanding of miRNA dysregulation in cancers.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal