HOXB4, a member of the Homeobox transcription factor family, promotes expansion of hematopoietic stem cells and hematopoietic progenitor cells in vivo and ex vivo when overexpressed. However, the molecular mechanisms underlying this effect are not well understood. To identify direct target genes of HOXB4 in primary murine hematopoietic progenitor cells, we induced HOXB4 function in lineage-negative murine bone marrow cells, using a tamoxifen-inducible HOXB4-ERT2 fusion protein. Using expression microarrays, 77 probe sets were identified with differentially changed expression in early response to HOXB4 induction. Among them, we show that Hemogen (Hemgn), encoding a hematopoietic-specific nuclear protein of unknown function, is a direct transcriptional target of HOXB4. We show that HOXB4 binds to the promoter region of Hemgn both ex vivo and in vivo. When we overexpressed Hemgn in bone marrow cells, we observed that Hemgn promoted cellular expansion in liquid cultures and increased self-renewal of myeloid colony-forming units in culture, partially recapitulating the effect of HOXB4 overexpression. Furthermore, down-regulation of Hemgn using an shRNA strategy proved that Hemgn contributes to HOXB4-mediated expansion in our myeloid progenitor assays. Our results identify a functionally relevant, direct transcriptional target of HOXB4 and identify other target genes that may also participate in the HOXB4 genetic network.
Introduction
Hematopoiesis arises from hematopoietic stem cells (HSCs), which have the unique capacity to generate multilineage blood cells and to self-renew to maintain the HSC pool.1 A delicate balance is maintained between different cellular fates of cycling HSCs: differentiation, self-renewal, and apoptosis.2 These developmental fates appear to be controlled both by extrinsic signals from the microenvironment and by intrinsic genetic programs,3 although the precise molecular mechanisms underlining these fates are still not comprehensively understood. Identifying molecules that promote HSC expansion is an important clinical goal because significant expansion of HSCs would allow many new transplantation-based therapies.4 However, ex vivo expansion of HSCs has been difficult to achieve and often results in differentiation and apoptosis.
Human HOXB4, a member of the Homeobox (HOX) transcription factor family, can promote HSC/hematopoietic progenitor cell (HPC) expansion both in vivo and ex vivo when ectopically expressed in murine bone marrow (BM) cells.5,6 HOXB4 overexpression also promotes specification of definitive HSCs from differentiating murine embryonic stem cells.7,8 Other HOX genes have been implicated in the regulation of normal and leukemic hematopoiesis.9 HOX family members share a common highly conserved homeodomain for binding to the transcriptional regulatory elements of target genes. The DNA-binding specificity, affinity, and transregulating potential of a specific HOX protein can be modified by several factors.10,11 HOXB4 regulates HSC/HPC expansion through a transcription-dependent mechanism and DNA binding is required for HSC expansion.12,13 These findings suggest that HOXB4 functions by binding directly to target genes and initiating a specific transcriptional program associated with self-renewal. The direct target genes by which HOXB4 promotes HSC/HPC expansion have not been identified. Recently, the tumor necrosis factor-α pathway has been shown to be downstream of HOXB414,15 ; however, it is unclear whether any of the genes in this pathway is a direct transcriptional target of HOXB4.
Here, we describe a short-term liquid culture system in which a tamoxifen (TAM)-regulated HOXB4-ERT2 fusion protein was overexpressed in primary murine lineage-negative (Lin−) BM cells, allowing us to temporally follow changes in gene expression after induction of HOXB4 function. We now report a set of gene expression changes that are modulated within 6 to 12 hours after HOXB4 induction. One particular target gene, Hemgn, which encodes a nuclear protein that is specifically expressed in hematopoietic tissues,16 was studied in further detail. We then developed a novel chromatin immunoprecipitation (ChIP) assay for HOXB4 to verify that the Hemgn promoter is a direct transcriptional target of HOXB4. Furthermore, we show that overexpression of Hemgn partially recapitulates the function of HOXB4 in promoting myeloid progenitor cells expansion ex vivo, in part by protecting BM cells from apoptosis during cell culture. Lastly, we prove that Hemgn plays a role in HOXB4-mediated expansion using an shRNA vector directed against Hemgn.
Methods
Vector construction and design
The human HOXB4 cDNA was kindly provided by Dr Keith Humphries (Terry Fox Laboratory, Vancouver, BC) and was directly used to generate the murine stem cell virus (MSCV)–HOXB4–internal ribosome entry sequence (IRES)–yellow fluorescent protein (YFP) vector. Polymerase chain reaction (PCR) cloning was used to generate the MSCV-HOXB4-ERT2-IRES–green fluorescent protein (GFP) vector by placing the sequence encoding the ERT2 estrogen receptor variant17,18 downstream of the HOXB4 cDNA. The PCR strategy generated a linker encoding the following sequence (CLQNSDQRNE) inserted immediately after the HOXB4 peptide sequence and upstream of the ERT2 sequence. The murine Hemgn cDNA (National Center for Biotechnology Information reference sequence NM_053149.2) was obtained from ATCC, sequenced, and inserted directly downstream of the long terminal repeat (LTR) promoter in the MSCV-IRES-GFP vector. A polyclonal population of ecotropic retroviral producer GPE+86 cells was generated, and retroviral vector supernatants freshly collected from the GPE+86 producer cells were produced as previously described.19
Separation, transduction, and analysis of BM cells
All animal experiments were done on protocols approved by the St Jude Institutional Animal Care and Use Committee. BM cells were harvested from the tibias and femurs of 7- to 12-week-old female C57BL/6J mice and were incubated with phycoerythrin (PE)-conjugated lineage antibody mixture (anti–mouse Mac-1, Gr-1, B220, CD4, CD8, Ter-119, BD Biosciences PharMingen). Lin− BM cells were enriched by depleting PE-labeled cells through anti-PE magnetic microbeads (Miltenyi Biotec) on autoMACS separator (Miltenyi Biotec) according to the manufacturer's instructions. The Lin−– or 5-fluorouracil (5-FU)–treated BM cells were prestimulated for 2 days with 20 ng/mL recombinant mouse interleukin-3 (IL-3), 50 ng/mL human IL-6, and 50 ng/mL mouse stem cell factor (R&D Systems) in Dulbecco modified Eagle medium (DMEM) supplemented with 15% fetal bovine serum (FBS), 100 units/mL penicillin, 100 μg/mL streptomycin, and 2mM l-glutamine (R&D Systems). The cells were then spin-infected at 500g for 30 minutes at room temperature using plates preloaded with 25 μg/mL RetroNectin (Takara) and retroviral vector supernatant at a multiplicity of infection of 10 to 20 transducing units per cell (tu/cell). After 3 or 4 rounds of transduction over 2 consecutive days, cells were collected, washed with phosphate-buffered saline (PBS), and then resuspended in cytokine-supplemented medium described above in this section.
TAM induction and myeloid progenitor assays
Cells were cultured in the presence of TAM before plating into semisolid methylcellulose medium for colony-forming units in culture (CFU-C) assay and before RNA extraction. A total of 10 mg of TAM was dissolved in 20 mL of distilled H2O to obtain a stock at 0.5 mg/mL. This stock solution was further diluted and directly added into the medium at a final concentration of 300nM. After 12, 24, and 48 hours of incubation, the cells were washed in PBS and then plated in methylcellulose medium, MethoCult GF M3434 (StemCell Technologies), for myeloid progenitor CFU-C assays. After 7 days, primary colonies were counted, collected, resuspended, and replated at 105 cells/mL in M3434 medium for an additional 10 days of culture. The resulting secondary colonies were then scored under a microscope.
Affymetrix RNA analysis
Total RNA was extracted from TAM-treated Lin− BM cells by RNA STAT-60 reagent (Tel-Test). RNA concentration and purity were determined by spectrophotometric analysis, and RNA integrity was confirmed by electrophoresis. Total RNA (100 ng) was processed according to the Affymetrix eukaryote 2-cycle target labeling protocol (http://www.affymetrix.com/support/technical/manual/expression_manual.affx). Biotin-labeled cRNA (20 μg) was hybridized overnight at 45°C to the Mouse-430v2 GeneChip array. After staining and washing, arrays were scanned and expression values were summarized using the MAS5 algorithm as implemented in the GCOS, Version 1.4 software (Affymetrix). Signals were normalized for each array by scaling to a 2% trimmed mean of 500. Detection calls (Present, Absent, and Marginal) were determined using the default parameters of the software. Signal values were log2-transformed before analysis. Differential expression between TAM-treated HOXB4-ERT2 and GFP-transduced cells was determined from 4 independent experiments using the paired t test (S-Plus, Version 6.2 software, TIBCO). Hierarchical clustering was performed using Spotfire Decision Site, Version 9.0 software (TIBCO).
Electrophoretic mobility shift assay and ChIP assays
A GST-tag was fused to the N-terminus of HOXB4 and the GST-HOXB4 fusion protein was expressed and purified using the pET-GST Fusion System 42 (Novagen, EMD Chemicals). Synthetic single-strand DNA oligos were biotin-labeled with biotin 3′ END DNA labeling kit (Pierce Chemical) and annealed. In supershift analysis, 1 μg anti-HOXB4 monoclonal antibody (clone I12; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA) was used. The sequence of the wild-type (WT) probe for the promoter of Hemgn is: 5′-ACACTCTGC[b]TAATTACAGCCTTT-3′ (−1562 bp to −1540 bp); and sequence of the mutant probe is: 5′-ACACTCTGC[b]AGCATACAGCCTTT-3′.
ChIP assays were performed using ChIP-IT Express kit (Active Motif). Briefly, BM cells transduced with the MSCV-HOXB4-IRES-YFP vector were fixed with 1% formaldehyde for 10 minutes at room temperature, and the chromatin from 2 × 107 cells was sonicated into fragments averaging approximately 500 bp. The chromatin-protein complex derived from 2.5 × 106 cells was immunoprecipitated with either 1 μg anti-HOXB4 antibody (I12) or 1 μg IgG2A isotype control antibody (R&D Systems). Immunoprecipitates were analyzed using semiquantitative PCR (Tag PCR core kit, QIAGEN) at the following parameters: 35 cycles with 94°C for 1 minute, 60°C for 1 minute, and 72°C for 1 minute. Quantitative real-time PCR was performed with SYBR Green DNA incorporation method using QuantiTect SBBR Green PCR kit (QIAGEN), carried out on StepOnePlus Real-Time PCR System (Applied Biosystems), and analyzed using the quantitative comparative CT (2−ΔΔCT) method using StepOne software, Version 2.0 (Applied Biosystems). The sequence of the PCR primers is provided in supplemental data (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Apoptosis and cell-cycle assays
Transduced BM cells were cultured in the cytokine-supplemented medium for 6 days, then washed with PBS, and recultured in medium containing 1% FBS and no cytokines. Sixteen hours later, 3 × 105 cells were stained with annexin V–allophycocyanin (BD Biosciences) and propidium iodide (PI). For cell-cycle analysis, the transduced cells were cultured for 6 days in the cytokine-supplemented medium and then were sorted to separate GFP/YFP-positive and -negative populations, which were, respectively, stained with PI for DNA content analysis.
shRNA knockdown of Hemgn
Four shRNA target sequences were designed using the Applied Biosystems siRNA target finder software (http://www.ambion.com/techlib/misc/siRNA_finder.html). The target sequence for the shRNA-6 vector was GCAGTTGAACCTGAATTCA and is located in the open reading frame of Hemgn mRNA (with GenBank accession no. NM_053149.2). A random, scrambled target sequence was used as a control. These sequences were ligated to pENTR/U6 vector (Invitrogen) downstream of U6 promoter and then inserted into cl20-MSCV-IRES-GFP lentiviral vector and verified by sequencing. The vector plasmid was used to generate lentiviral vector supernatants using transient transfection of vector and helper plasmids as previously described.20
Mouse erythroleukemia (MEL) cells express Hemgn endogenously and were used to verify activity of the shRNA vectors. MEL cells were transduced with each of the shRNA lentiviral vectors. Forty-eight hours later, total protein was harvested from cell lysates and analyzed by Western blot using the M-180 polyclonal rabbit antibody (Santa Cruz Biotechnology) at a dilution of 1:2000. For the BM cell experiments, harvested cells were first transduced with the MSCV-HOXB4-IRES-YFP vector. Forty-eight hours later, YFP-positive cells were sorted and then transduced with shRNA-GFP lentiviral particles for another 48 hours. Flow cytometric sorting was used to isolate GFP/YFP double-positive cells for use in subsequent assays.
Results
Inducible HOXB4 activity ex vivo
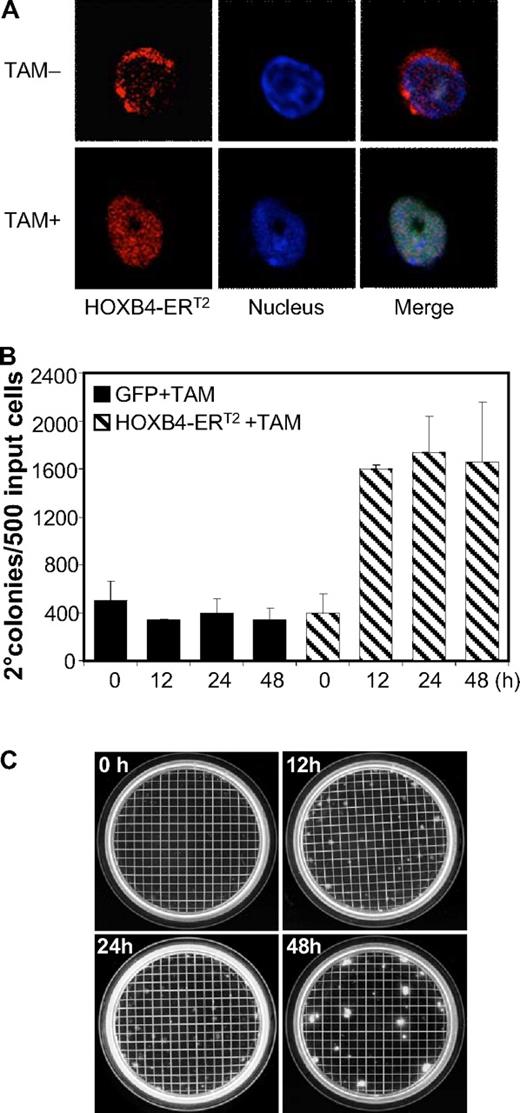
To identify potential direct target genes of HOXB4 in murine BM cells, a HOXB4-ERT2 fusion gene was developed to allow regulated HOXB4 activity by TAM (supplemental Figure 1A). A retroviral vector expressing this fusion protein under the control of the MSCV promoter was generated, and expression of HOXB4-ERT2 protein was verified in the vector producer cells (supplemental Figure 1B). TAM-induced nuclear translocation of HOXB4-ERT2 in the transduced BM cells was confirmed by immunofluorescence staining (Figure 1A).
Effect of TAM-induced HOXB4-ERT2 activity on myeloid CFU-C. (A) TAM-induced cytosol to nucleus translocation of HOXB4-ERT2 in Lin− BM cells confirmed by confocal microscopy (red represents HOXB4-ERT2; and blue, nucleus dye). Slides were imaged with a Nikon TE2000-E inverted microscope equipped with a CISi confocal system with EZCl software and an 40×/1.3 NA Plan Apo objective. (B) TAM treatment for 12, 24, and 48 hours induced an increase in secondary myeloid progenitor colony formation in HOXB4-ERT2-overexpressing Lin− BM cells ex vivo. (C) Pictures of the secondary myeloid progenitor colony plates from HOXB4-ERT2-overexpressing cells treated with TAM for 0, 12, 24, and 48 hours.
Effect of TAM-induced HOXB4-ERT2 activity on myeloid CFU-C. (A) TAM-induced cytosol to nucleus translocation of HOXB4-ERT2 in Lin− BM cells confirmed by confocal microscopy (red represents HOXB4-ERT2; and blue, nucleus dye). Slides were imaged with a Nikon TE2000-E inverted microscope equipped with a CISi confocal system with EZCl software and an 40×/1.3 NA Plan Apo objective. (B) TAM treatment for 12, 24, and 48 hours induced an increase in secondary myeloid progenitor colony formation in HOXB4-ERT2-overexpressing Lin− BM cells ex vivo. (C) Pictures of the secondary myeloid progenitor colony plates from HOXB4-ERT2-overexpressing cells treated with TAM for 0, 12, 24, and 48 hours.
To test whether this construct led to Tam-induced expansion of myeloid progenitors, an established secondary CFU-C assay was used as previously described.5 The transduced Lin− cells (60%-70% transduction efficiency in multiple experiments) were cultured in 300nM TAM for 0, 12, 24, and 48 hours, washed, and then plated into semisolid methylcellulose medium for myeloid progenitor CFU-C assay. After 7 days, cells were harvested, resuspended, and plated for secondary CFU-C formation, a function associated with HOXB4-induced self-renewal. TAM treatment for 12 hours or greater resulted in an increase in secondary CFU-C relative both to TAM-treated GFP control cells and to HOXB4-ERT2 cells in the absence of TAM (Figure 1B-C). At 48 hours, we saw an increase in size of colonies as well. These results demonstrate that the HOXB4-ERT2 fusion protein functions in a TAM-dependent manner and verifies that the system does not have leaky function in the absence of TAM.
Identification and validation of direct HOXB4 target genes
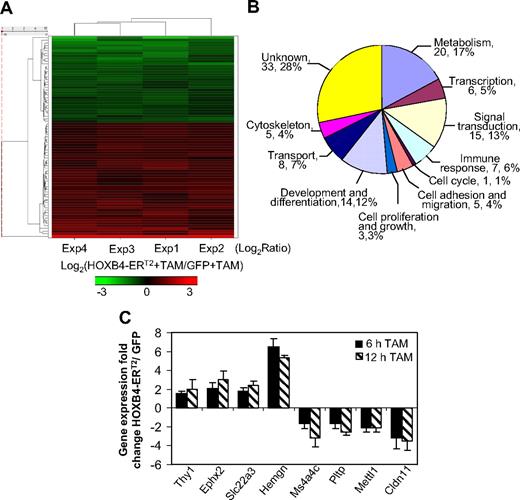
To identify potential direct target genes of HOXB4 involved in expansion of myeloid progenitors, RNA was extracted 12 hours after TAM treatment of transduced, Lin− BM cells and was analyzed for differential gene expression change using Affymetrix 430v2 chips. The transduction efficiency ranged from 60% to 70% in multiple experiments. Analysis of 4 independent experiments showed good reproducibility in gene expression changes. The Pearson correlation coefficient between all experimental samples ranged from 0.91 to 0.95 among probe sets with more than one “Present” call (supplemental Table 1), indicating that there existed low experimental variation and that HOXB4 overexpression did not cause a global change in gene expression. First, we manually excluded those probe sets showing more than 4 “Absent” calls in a total of 8 samples from the 4 experiments. A probe set was considered as a potential downstream target of HOXB4 if the difference between the mean expression values (HOXB4-ERT2 vs GFP) was at least 1.5-fold change, and the extent of the expression difference was statistically significant (P < .01) as evaluated by paired t test. We chose a 1.5-fold threshold because 30% to 40% of the cells in a given sample were not transduced; hence, the 1.5-fold change measured from the samples would underestimate what actually occurred in the transduced cells.
A total of 77 probe sets were identified as significantly changed by this analysis. Among them, 44 were up-regulated (supplemental Table 2) and 33 were down-regulated (supplemental Table 3) by HOXB4 activation. A hierarchical clustering analysis and heat map were generated for those 77 probe sets from the 4 independent experiments (Figure 2A). Functional classification of those probe sets by Gene Ontology annotations denoting different biologic processes (Figure 2B) showed a relatively high percentage of probe sets involved in unknown (n = 33, 28%), metabolism (n = 20, 17%), signal pathways (n = 15, 13%), and development and differentiation (n = 14, 12%).
Identification of candidate HOXB4 target genes and validation of their expression. (A) Hierarchical cluster diagram and heat map of the 77 candidate probe sets selected from 4 biologically independent experiments using the log2-ratio of TAM-induced HOXB4-ERT2 versus TAM-induced GFP expression values observed within each of the 4 experiments. (B) Functional classification of the 77 probe sets using Gene Ontology annotations. Some probe sets were classified into more than 1 category; thus, the sum of all 11 groups is more than 77. (C) Quantitative real-time PCR confirmation of the expression change of the selected 8 candidate target genes from independent experiments with TAM treatment for 6 hours or 12 hours in HOXB4-ERT2 vector-transduced and GFP control-transduced Lin− BM cells.
Identification of candidate HOXB4 target genes and validation of their expression. (A) Hierarchical cluster diagram and heat map of the 77 candidate probe sets selected from 4 biologically independent experiments using the log2-ratio of TAM-induced HOXB4-ERT2 versus TAM-induced GFP expression values observed within each of the 4 experiments. (B) Functional classification of the 77 probe sets using Gene Ontology annotations. Some probe sets were classified into more than 1 category; thus, the sum of all 11 groups is more than 77. (C) Quantitative real-time PCR confirmation of the expression change of the selected 8 candidate target genes from independent experiments with TAM treatment for 6 hours or 12 hours in HOXB4-ERT2 vector-transduced and GFP control-transduced Lin− BM cells.
To validate the expression data from this microarray assay, 8 genes that showed relatively large changes in expression were chosen for quantitative real-time PCR confirmation (Figure 2C). The housekeeping gene Gapdh was used as an internal control for gene expression normalization. This assay validated the microarray data and also showed that changes in the gene expression present 12 hours after TAM treatment were also present 6 hours after TAM treatment (Figure 2C), reflecting early transcriptional events after HOXB4 activation.
Direct activation of Hemgn by HOXB4
Hemgn was one of the genes that was up-regulated at a relatively high level and appeared to us to be a potentially interesting candidate gene. Hemgn is a nuclear protein of unknown function that is specifically expressed in undifferentiated hematopoietic cells,21 and overexpression of Hemgn in cell lines inhibits apoptosis and differentiation,22 consistent with a potential effect in self-renewal processes. Overexpression of Hemgn in a transgenic mouse model caused an expansion of myeloid hematopoiesis, most particularly in the spleen,23 and overexpression of the human homolog, EDAG, has been found in acute human leukemia.24 Based on these findings, we decided to pursue the role of Hemgn in mediating the HOXB4 phenotype.
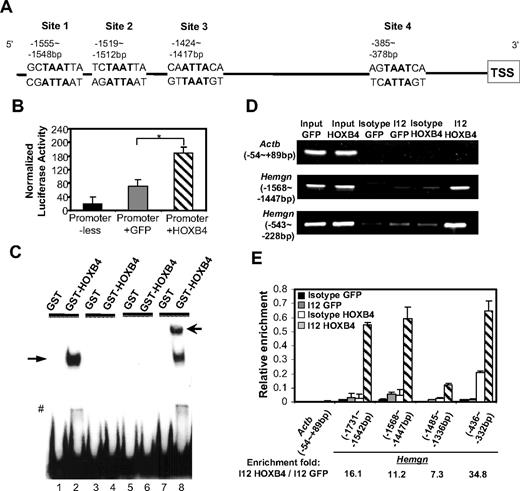
Inspection of the promoter sequence of Hemgn reveals 4 putative HOXB4-binding sites (Figure 3A) with a “TAAT” core and HOXB4-preferred flanking nucleotides. The −1887 bp to 213 bp region containing the 4 putative HOXB4-binding sites and the transcription start site was cloned upstream of a luciferase reporter gene, and this construct was tested in a transient transfection assay in MEL cells. When this construct was cotransfected with an internal Renilla control vector and the MSCV vector expressing either HOXB4 or GFP control, HOXB4 significantly increased the luciferase activity in MEL cells (Figure 3B), indicating that the promoter of Hemgn can be directly activated by HOXB4.
HOXB4 transcriptionally activates and binds to the promoter of Hemgn bothex vivoand in vivo. (A) Schematic illustration of the multiple putative HOXB4-binding sites on the promoter region of Hemgn (sites 1, 2, 3, and 4). (B) HOXB4-dependent increase in promoter activity of Hemgn-luciferase in MEL cell cotransfection experiments. *P < .05. (C) HOXB4-specific binding caused band shift of probes containing the putative HOXB4-binding site (site 1). Lanes 1 and 2 indicate biotin-labeled WT probe (−1562 bp to −1540 bp of Hemgn promoter); lanes 3 and 4, labeled WT probe: competitive unlabeled probe = 1:200; lanes 5 and 6, probe with “TAAT” core sequence mutated to “AGCA”; lanes 7 and 8, labeled WT probe plus anti HOXB4 antibody (I12); solid arrow indicates band shift by HOXB4; and open arrow, supershift by I12. #Degraded GST-HOXB4 caused band shift. (D) Semiquantitative PCR detected enrichment of the promoter fragments of Hemgn by HOXB4 specific binding after ChIP assay in 5-FU BM cells. (E) Quantitative real-time PCR detected relative enrichment of individual regions in Hemgn promoter by HOXB4 after ChIP assay in 5-FU BM cells. All values were normalized to the corresponding input control sample.
HOXB4 transcriptionally activates and binds to the promoter of Hemgn bothex vivoand in vivo. (A) Schematic illustration of the multiple putative HOXB4-binding sites on the promoter region of Hemgn (sites 1, 2, 3, and 4). (B) HOXB4-dependent increase in promoter activity of Hemgn-luciferase in MEL cell cotransfection experiments. *P < .05. (C) HOXB4-specific binding caused band shift of probes containing the putative HOXB4-binding site (site 1). Lanes 1 and 2 indicate biotin-labeled WT probe (−1562 bp to −1540 bp of Hemgn promoter); lanes 3 and 4, labeled WT probe: competitive unlabeled probe = 1:200; lanes 5 and 6, probe with “TAAT” core sequence mutated to “AGCA”; lanes 7 and 8, labeled WT probe plus anti HOXB4 antibody (I12); solid arrow indicates band shift by HOXB4; and open arrow, supershift by I12. #Degraded GST-HOXB4 caused band shift. (D) Semiquantitative PCR detected enrichment of the promoter fragments of Hemgn by HOXB4 specific binding after ChIP assay in 5-FU BM cells. (E) Quantitative real-time PCR detected relative enrichment of individual regions in Hemgn promoter by HOXB4 after ChIP assay in 5-FU BM cells. All values were normalized to the corresponding input control sample.
Among the 4 putative HOXB4-binding sites (Figure 3A), site 1 (−1555 bp to −1548 bp) contains the most favored flanking nucleotides for high-affinity HOXB4 binding. To directly test whether HOXB4 can bind to this site, we designed DNA probes spanning this specific site (−1562 bp to −1540 bp) to examine the binding of HOXB4 using electrophoretic mobility shift assay. These experiments showed that HOXB4 binding caused a band shift that was detected when the biotin-labeled WT probe was incubated with purified GST-HOXB4 protein (Figure 3C lane 2). No band shift was detected in the reactions with a competitive unlabeled probe (lane 4), or a probe containing mutated “TAAT” core sequence (lane 6), or with control GST protein (lanes 1, 3, 5, and 7). Addition of a HOXB4 antibody resulted in supershift of the specifically retarded fragment (lane 8). These results demonstrate that HOXB4 can directly bind to site 1 in the promoter region of Hemgn in vitro.
To discern whether HOXB4 is recruited to the promoter of Hemgn in BM cells in vivo, we developed a ChIP assay for HOXB4 binding using the I12 anti-HOXB4 antibody that efficiently immunoprecipitated HOXB4 from a transduced cell line (supplemental Figure 2). We designed semiquantitative PCR primer pairs to span the −1568 bp to −1447 bp region containing 2 putative HOXB4-binding sites (sites 1 and 2) or to span the −543 bp to −228 bp region, which contains one putative site (site 4).
BM cells transduced with either HOXB4 or GFP control vectors were used in this ChIP assay, and the immunoprecipitated DNA/chromatin fragments were released for PCR analysis. Using HOXB4-transduced cells, both binding site regions were specifically enriched in immunoprecipitation reactions using the I12 anti-HOXB4 antibody (Figure 3D). In contrast, neither of the 2 regions was enriched in cells transduced with the GFP control vector or in HOXB4 vector-transduced cells using an isotype control antibody. These results verify that HOXB4 binds to the promoter of Hemgn in vivo in primary BM cells overexpressing HOXB4.
To further quantify HOXB4 binding at the individual “TAAT” sites, quantitative real-time PCR primers were designed to amplify the individual putative HOXB4-binding sites in the promoter of Hemgn (Figure 3E). The −1731 bp to −1542 bp (site 1) and the −436 bp to −332 bp (site 4) regions were enriched at relatively higher levels (16.1-fold and 34.8-fold). As sites 1, 2, and 3 are relatively close to each other and because the sonicated DNA fragments average approximately 500 bp in length, it is possible that binding to sites 1, 2, and 3 is not necessarily an independent event in this assay.
Overexpression of Hemgn leads to expansion of BM cells in culture
We next tested whether overexpression of Hemgn could contribute to the expansion phenotype of HOXB4. A full-length cDNA of murine Hemgn was cloned into the MSCV-IRES-GFP retroviral vector, and expression of Hemgn was confirmed in GPE+86 vector producer cells by Western blot analysis (supplemental Figure 3). Because the Hemgn vector had a relatively low titer (5 × 104 tu/mL), the transduction efficiency using the MSCV-Hemgn-IRES-GFP vector was only approximately 10% to 15% for mouse BM cells by flow cytometry. Therefore, we diluted the GFP control and HOXB4-transduced BM cells with mock-transduced cells to the same proportion in subsequent liquid culture and CFU-C experiments.
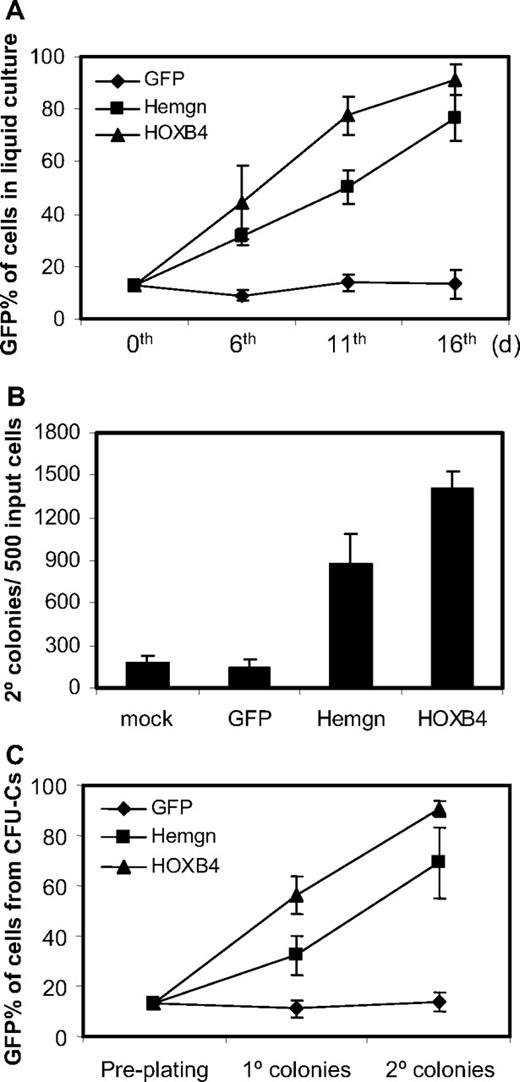
Cells transduced either with the MSCV-Hemgn-IRES-GFP vector or with the MSCV-HOXB4-IRES-YFP vector had a significant growth advantage over nontransduced cells in liquid culture (Figure 4A). This is demonstrated by a progressive increase in the GFP percentage in Hemgn-overexpressing cells and YFP percentage in HOXB4-overexpressing cells over the 16-day culture period (Figure 4A). In contrast, the cells transduced with GFP control vector did not show any growth advantage during the culture time.
Growth advantage of Hemgn-overexpressing BM cells in culture and in serial CFU-C assays. (A) Flow cytometry measurements of the GFP/YFP percentages of transduced BM cells at 6th, 11th, and 16th day in liquid culture after transduction. Note that on day 0, dilutions were performed so that all samples had 13% transduced cells dilute with mock-transduced cells. (B) Secondary CFU-C colony numbers generated the transduced cells. The vector groups are shown below each histogram. (C) The GFP/YFP percentage of transduced cells (GFP/YFP) in primary and secondary CFU-C colonies, which were derived from the preplating 13% GFP/YFP-positive cell mixture. Three biologically independent experiments were performed, and the SD is shown in each panel.
Growth advantage of Hemgn-overexpressing BM cells in culture and in serial CFU-C assays. (A) Flow cytometry measurements of the GFP/YFP percentages of transduced BM cells at 6th, 11th, and 16th day in liquid culture after transduction. Note that on day 0, dilutions were performed so that all samples had 13% transduced cells dilute with mock-transduced cells. (B) Secondary CFU-C colony numbers generated the transduced cells. The vector groups are shown below each histogram. (C) The GFP/YFP percentage of transduced cells (GFP/YFP) in primary and secondary CFU-C colonies, which were derived from the preplating 13% GFP/YFP-positive cell mixture. Three biologically independent experiments were performed, and the SD is shown in each panel.
We then examined whether Hemgn would have a similar effect to HOXB4 in the CFU-C secondary replating assay. Equivalent mixtures of transduced cells were plated in the methylcellulose medium and replated 7 days later for generating secondary CFU-C colonies. We found that Hemgn-overexpressing BM cells generated more secondary CFU-C colonies than mock (4.9-fold) and that of GFP control cells (6.1-fold; Figure 4B). As a positive control, the HOXB4-overexpressing BM cells generated 7.9-fold and 9.9-fold more secondary colonies than mock and the GFP control cells, respectively. The proportion of GFP/YFP-positive cells was scored by flow cytometry both from primary and secondary CFU-C dishes and showed that, with Hemgn-transduced cells, the GFP percentage increased to 32% and 69% in cells collected from primary and secondary colonies, respectively (Figure 4C). As a positive control, YFP percentage in HOXB4-overexpressing cells increased to 58% and 91% in primary and secondary colony cells, respectively. The GFP control BM cells did not show any expansion and were maintained at approximately 13% transduced cells. These results show that Hemgn and HOXB4 both promoted myeloid progenitor cells to expand both in liquid culture and in semisolid CFU-Cs.
Hemgn and HOXB4 protect BM cells from apoptosis
To explore the cellular mechanism by which Hemgn overexpression leads to expansion, BM cells from 5-FU–treated mice were transduced and then cultured for 6 days in DMEM medium containing 15% FBS, IL-3, IL-6, and stem cell factor. We then transferred these cells into DMEM medium containing 1% FBS and no cytokines for 16 hours to induce apoptosis. These cultures were then analyzed by gating on transduced cells and assaying for induction of apoptosis by staining with annexin V and PI, with the GFP- or YFP-negative populations serving as negative internal controls.
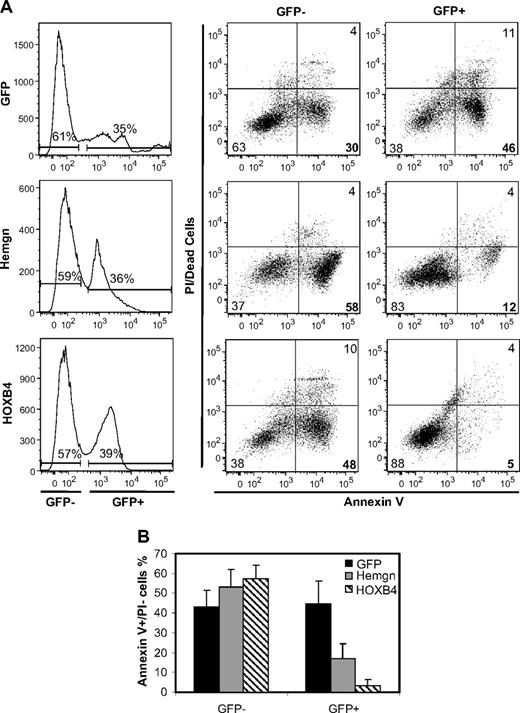
A representative experiment (Figure 5A) shows that both Hemgn and HOXB4 vector-transduced GFP-positive cells had a much smaller fraction of annexin V+/PI− cells than nontransduced cells or GFP control-transduced cells. Three independent experiments were performed and showed that both HOXB4 and Hemgn protected transduced cells from apoptosis associated with serum and cytokine deprivation, although HOXB4 was more potent in this regard (Figure 5B).
Inhibition of apoptosis in cells transduced with the Hemgn or the HOXB4 vector. (A) A representative experiment in which primary murine bone marrow cells were transduced with the indicated vectors, grown for 6 days in complete medium, washed, and then cultured for 16 hours in serum-deprived medium without cytokines to induce apoptosis. (Left panel) Gating for transduced cells (GFP/YFP+). The gated nontransduced cells serve as internal negative controls for transduction. Staining within each population for annexin V and PI is shown in middle and right column. (Middle column) Results for the nontransduced cells. (Right column) Gated, transduced cells. The percentage of apoptotic cells is indicated in the lower right quadrant for each sample. (B) Quantification and statistics of 3 biologically independent experiments showing the proportion of apoptotic cells for the 3 vectors and for the control, nontransduced cells in each sample.
Inhibition of apoptosis in cells transduced with the Hemgn or the HOXB4 vector. (A) A representative experiment in which primary murine bone marrow cells were transduced with the indicated vectors, grown for 6 days in complete medium, washed, and then cultured for 16 hours in serum-deprived medium without cytokines to induce apoptosis. (Left panel) Gating for transduced cells (GFP/YFP+). The gated nontransduced cells serve as internal negative controls for transduction. Staining within each population for annexin V and PI is shown in middle and right column. (Middle column) Results for the nontransduced cells. (Right column) Gated, transduced cells. The percentage of apoptotic cells is indicated in the lower right quadrant for each sample. (B) Quantification and statistics of 3 biologically independent experiments showing the proportion of apoptotic cells for the 3 vectors and for the control, nontransduced cells in each sample.
Hemgn did not have any direct effects on proliferation or cell-cycle status. In complete media, the percentage of cells at the G1, S, and G2/M phases of the cell cycle was not altered by Hemgn overexpression (supplemental Figure 4). Cells transduced with the HOXB4 vector had a slight increase in percentage in S-phase compared with control (49% and 42%, respectively), suggesting that HOXB4, unlike Hemgn, could have modest effects on proliferation in this system.
Hemgn is required for full HOXB4-mediated expansion
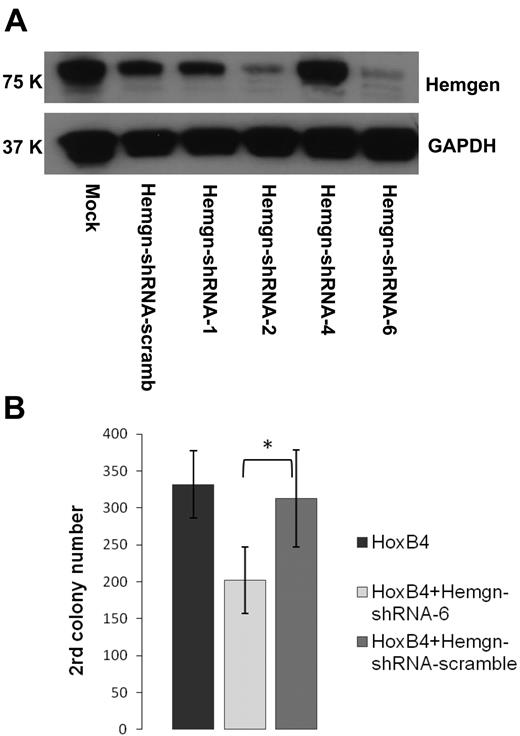
To determine whether induction of Hemgn by HOXB4 was required for expansion of myeloid progenitors, we knocked down Hemgn expression in HOXB4-transduced cells using shRNA technology. We designed 4 Hemgn-shRNA lentiviral constructs that coexpress a linked GFP gene and tested their function in MEL cells, which express Hemgn endogenously and at a relatively high level. The shRNA 6 construct gave the highest degree of inhibition, reducing Hemgn protein levels to approximately 20% of endogenous levels seen in mock controls (Figure 6A). Hemgn-shRNA-6 targets the coding sequence of the Hemgn mRNA at 1075 to 1093 bp downstream of the start codon.
Knockdown of Hemgn by shRNA decreased secondary CFU-C in HOXB4-transduced BM cells. (A) Western blot analysis for Hemgn expression in MEL cells transduced with panel of shRNA lentiviral vectors. A scrambled control sequence was included as well as mock-transduced cells. shRNA-6 was the most effective construct and used in the CFU-C experiments shown. (B) In these experiments, HOXB4-transduced cells were then transduced with either the shRNA-6 or the scrambled control vector and assayed in a secondary CFU-C assay. The secondary colony number per 104 plated cells is shown for HOXB4 only, for HOXB4 transduced with shRNA-6, and for the scrambled control. Three independent experiments were performed, and the decrease in secondary colony number with the shRNA was highly significant. *P < .05.
Knockdown of Hemgn by shRNA decreased secondary CFU-C in HOXB4-transduced BM cells. (A) Western blot analysis for Hemgn expression in MEL cells transduced with panel of shRNA lentiviral vectors. A scrambled control sequence was included as well as mock-transduced cells. shRNA-6 was the most effective construct and used in the CFU-C experiments shown. (B) In these experiments, HOXB4-transduced cells were then transduced with either the shRNA-6 or the scrambled control vector and assayed in a secondary CFU-C assay. The secondary colony number per 104 plated cells is shown for HOXB4 only, for HOXB4 transduced with shRNA-6, and for the scrambled control. Three independent experiments were performed, and the decrease in secondary colony number with the shRNA was highly significant. *P < .05.
We then used this vector to cotransduce BM cells that had also been transduced with the MSCV-HOXB4-IRES-YFP construct. Flow cytometry was then used to isolate doubly transduced BM cells expressing both the GFP and YFP reporter genes. These cotransduced cells were then evaluated in the secondary CFU-C replating assay in 3 independent experiments. The number of secondary CFU-C was significantly decreased in cells transduced with the Hemgn knockdown vector (Figure 6B). An approximate 40% reduction in colony formation was noted both comparison with HOXB4-transduced cells and with cells transduced both with HOXB4 and with a random shRNA control vector (P < .05; Figure 6B). These results show that Hemgn plays at least a partial functional role in HOXB4-mediated expansion. The fact that secondary colony formation was not reduced to levels seen in the absence of HOXB4 may be explained by the lack of complete Hemgn knockdown with the shRNA-6 construct and/or the possibility that other genes in the HOXB4 network can independently contribute to the expansion phenotype.
Discussion
Because HOX proteins act as transcription factors, identifying their downstream target genes is critical for elucidating the molecular mechanisms of their various functions. Gene expression profiling analysis has been used for this purpose; however, only a small number of HOX targets have yet been identified.25 Specifically, little is known about the direct target genes of HOXB4 that mediate the expansion of HSCs and HPCs. We have used an inducible HOXB4-ERT2 system to identify direct targets of HOXB4 in primary murine, Lin− BM cells. Our studies have identified 77 potential targets of HOXB4 in this progenitor-enriched population of primary hematopoietic cells, including 15 genes involved in signal transduction, 14 genes involved in other aspects of development and differentiation, 6 genes that are transcription factors, and 33 genes of unknown function. Although we have focused on Hemgn as a specific candidate, it is probable that multiple other genes identified in our array studies have important roles in the expansion phenotype and that this snapshot of the HOXB4 transcriptional network portrays what is probably a complex, multifunctional genetic program. For example, our finding that HOXB4 but not Hemgn can cause increased proliferation suggests that cell cycle targets could cooperate with the antiapoptotic effects of Hemgn to induce expansion.
Our screen was designed to detect the genes activated by overexpression of HOXB4 and therefore participate in exogenously induced, self-renewal program. We recognize that these data may not necessarily identify target genes for endogenously expressed Hoxb4, a setting where Hoxb4 is expressed at much lower levels. Hoxb4 and its other 3 paralogs, Hoxa4, Hoxc4, and Hoxd4, have a similar effect in promoting HPC expansion ex vivo.26,27 This redundancy may explain the relatively mild hematopoietic phenotype in Hoxb4−/− mice,28 and it is possible that group 4 paralogs share common downstream effectors for inducing expansion in HSCs/HPCs. In this regard, we performed quantitative real-time reverse-transcribed PCR assay in Hoxb4-deficient cells from the knockout mice and detected a 5-fold decrease in Hemgn mRNA expression in KSL cells compared with WT mice (supplemental Figure 5). These results indicate that Hemgn may also be regulated by endogenous Hoxb4 but that Hemgn expression is still expressed at a lower level in the absence of Hoxb4, possibly because of expression of functional paralogs.
We decided to focus on Hemgn, based on its relatively high degree of induction, its relatively specific hematopoietic expression pattern, and the paucity of knowledge regarding the function of this gene and its potential role in hematopoiesis. Hemgn is a recently discovered nuclear protein that is restrictively expressed in testis and hematopoietic tissues.16,21 It shows a spatial and temporal expression pattern in hematopoietic sites during embryogenesis and is expressed in blood islands of yolk sac, then in fetal liver, and finally in the adult spleen and BM.21 In adult BM, Hemgn is predominantly expressed in primitive progenitor and stem cell populations, but not in terminal differentiated mature blood cells. Human HEMGN, also called EDAG, shows a similar expression pattern in BM cells and, interestingly, maps to chromosome 9q22, which contains well-known leukemic breakpoints.21 Relatively high levels of HEMGN expression have been observed in human acute myeloid leukemia BM cells.24 In transgenic mice in which HEMGN was driven by human CD11a promoter, enhancement of myelopoiesis and suppression of lymphopoiesis were found, suggesting its regulatory role in hematopoiesis.23 The mechanism of his effect may be mediated by nuclear factor-κB.22 These facts suggest that Hemgn could play an important role as a hematopoietic regulator.
Our first goal was to determine whether Hemgn was a direct transcriptional target for HOXB4. We developed a novel in vivo HOXB4-binding assay based on chromatin immunoprecipitation and were able to verify HOXB4 binding to the Hemgn promoter in at least 2 different binding sites within primary BM cells transduced with the HOXB4 vector. This assay should prove useful in identifying other direct binding sites for HOXB4, potentially at a whole genome level. We next found that overexpression of Hemgn in murine BM cells leads to a partial replication of the HOXB4 phenotype, with significant expansion of transduced cells in liquid cultures and increased CFU-C self-renewal in secondary replating assays. These studies show that overexpression of Hemgn can confer a HOXB4-like phenotype with regard to HPC expansion. Further evidence was obtained to show that induction of Hemgn expression is necessary for the full HOXB4 phenotype. These experiments used an shRNA vector that partially knocks down Hemgn expression. When this vector was introduced into HOXB4-transduced cells, the self-renewal of myeloid progenitor cells was significantly attenuated. The fact that expansion was only partially inhibited is consistent with a necessary role of other HOXB4 target genes in the network, the fact that the shRNA knockdown was only partial, or both.
An important question is how Hemgn promotes expansion of HPCs. A large body of evidence suggests that suppression of apoptosis is required for HSC survival and self-renewal.29,–31 HEMGN has been reported to enhance survival of Ba/F3 cells by protecting them from apoptosis after IL-3 withdrawal.22 HOXB4 has also been shown to protect BM cells from the apoptosis-inducing effects of the tumor necrosis factor-α pathways.14 Our own data in primary mouse BM cells show that Hemgn protects cells from apoptosis because of serum and cytokine withdrawal.
It is probable that expansion of HSC/HPCs requires the combination of both survival and proliferation signals together with the inhibition of apoptosis and differentiation programs; and in this regard, Hemgn may also function by inhibiting hematopoietic differentiation. HEMGN overexpression can block the differentiation of HL-60 cells,22 which may be contributing to the partial block in myeloid differentiation seen with high levels of ectopic HOXB4 expression.32,33 Inspection of the peptide sequence for Hemgn reveals a nuclear localization motif and a coiled-coiled domain, the latter domain being associated with protein-protein interactions. Therefore, it is possible that Hemgn interacts with other transcription factors and modulates expression of genes involved in apoptosis. The physiologic function of Hemgn is unknown, and there are as yet no reports of a Hemgn null animal model, which will be an important experiment to more fully understand the role of this protein in steady-state and stress hematopoiesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Note added in proof:
In this issue of Blood (2010;116(5):720-730), Lee et al describe their study of HOXB4 target genes in a murine hematopoietic progenitor cell line, EML. We note that there appears to be little overlap with the genes we have identified in our study. We believe this can be explained in part by the different cellular systems used (EML cells vs Lin− bone marrow cells). For example, Lee et al identified B220 as a target gene that we did not see and would not expect to see in our B220-depleted, Lin− population. Lee et al also report up-regulation of Laptm4b and Gp49a expression by HOXB4 in LSK EML cells. Although we did not see these genes in our study, preliminary work in our laboratory has also shown up-regulation of both Laptm4b and Gp49a when Tam was used to induce HOXB4 in primitive bone marrow cells from transplanted mice (Y.S., unpublished data). In another experiment, we have found that HOXB4 can induce the Laptm4b promoter in a transient luciferase assay done in 293T cells and that the promoter for Laptm4b was enriched 2-fold in a HOXB4 chromatin immunoprecipation assays done using 5FU bone marrow cells (J.J., unpublished data). Therefore, our lack of detection of these events in Lin− cells grown in vitro likely reflects methodological differences rather than a lack of significance. Altogether, we believe the results from Lee et al complement our study and show the important influence of the cellular context for identifying HOXB4 target genes.
Acknowledgments
The authors thank Dr Richard Ashmun and the Flow Cytometry Core assistance in all the flow cytometry experiments, Hartwell Center for Bioinformatics and Biotechnology for Affymetrix chips hybridization and analysis, Cell and Tissue Imaging Center for the confocal imaging, and Dr Ji Zhang for invaluable scientific suggestions and technique help.
This work was supported by the National Heart, Lung, and Blood Institute (program project grant HL53749), the Assisi Foundation of Memphis, Cancer Center (support grant P30 CA 21765), and the American Lebanese Syrian Associated Charities.
National Institutes of Health
Authorship
Contribution: J.J. designed and performed the research, analyzed data, and wrote the paper; H.Y., Y.S., and T.L. performed research; G.N. and S.Z. analyzed data; and B.P.S. designed the research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Brian P. Sorrentino, Experimental Hematology Division, Hematology Department, MS 341, Rm D-2038F, Thomas Tower, St Jude Children's Research Hospital, 262 Danny Thomas Pl, Memphis, TN 38105-3678; e-mail: brian.sorrentino@stjude.org.