Abstract
The recent discovery of microRNAs (miRNAs) has revealed a new layer of gene expression regulation, affecting the immune system. Here, we identify their roles in regulating human plasmacytoid dendritic cell (PDC) activation. miRNA profiling showed the significantly differential expression of 19 miRNAs in PDCs after Toll-like receptor 7 (TLR7) stimulation, among which miR-155* and miR-155 were the most highly induced. Although they were processed from a single precursor and were both induced by TLR7 through the c-Jun N-terminal kinase pathway, miR-155* and miR-155 had opposite effects on the regulation of type I interferon production by PDC. Further study indicated that miR-155* augmented interferon-α/β expression by suppressing IRAKM, whereas miR-155 inhibited their expression by targeting TAB2. Kinetic analysis of miR-155* and miR-155 induction revealed that miR-155* was mainly induced in the early stage of stimulation, and that miR-155 was mainly induced in the later stage, suggesting their cooperative involvement in PDC activation. Finally, we demonstrated that miR-155* and miR-155 were inversely regulated by autocrine/paracrine type I interferon and TLR7-activated KHSRP at the posttranscriptional level, which led to their different dynamic induction by TLR7. Thus, our study identified and validated novel miRNA-protein networks involved in regulating PDC activation.
Introduction
Plasmacytoid dendritic cell (PDC) is a distinct dendritic cell type, specialized for the rapid secretion of type I interferon (type I IFN) in response to viruses.1-3 It has been demonstrated that PDCs can coordinate events during the course of viral infection, autoimmune diseases, and cancer. PDCs, through their production of interferon-α (IFN-α) and other cytokines, and through antigen presentation, link the innate and adaptive immune responses.3 PDC deficiency, leading to low levels of IFN-α production, results in an inadequate immune response, entailing susceptibility to viral infections or cancer, whereas excessive secretion of IFN-α can induce hyperimmune activation, which may lead to autoimmune disease or, in the case of HIV infection, CD4+ T-cell death.2-5 Therefore, type I IFN production by PDCs must be under tight control to prevent improper immune responses, which could be harmful to the host.2,3
PDCs express high levels of Toll-like receptor 7 (TLR7) and TLR9. The interaction between TLR7/9 and their ligands leads to the activation of the myeloid differentiation primary response gene 88/IL-1/4 receptor-associated kinase/tumor necrosis factor (TNF) receptor-associated factor 6/IκB kinases (MyD88/IRAK1/4/TRAF6/IKKs) pathway and the subsequent phosphorylation of interferon-regulatory receptor 7 (IRF-7), which is translocated into the nucleus and initiates IFN-α transcription.2,6 The phosphatidylinositol 3-kinase/Av-akt murine thymoma viral oncogene homolog 1/mammalian target of rapamycin (PI3K/AKT/mTOR) pathway and p38 mitogen-activated protein kinase (MAPK) activity have also been shown to positively regulate type I IFN production.7,8 In contrast to these positive regulators, an array of surface receptors on PDCs, such as blood dendritic cell antigen 2 (BDCA2), dendritic cell immunoreceptor (DCIR), immunoglobulin-like transcript 7 (ILT7), high-affinity immunoglobulin E receptor (FcϵRI), and natural killer partner 44 (NKp44), are reported to signal through a powerful immunoreceptor tyrosine-based activation motif (ITAM)–mediated, B-cell receptor (BCR)–like regulatory pathway to counter-regulate the prominent TLR signaling pathway.2,9-13 Although the kinetics of type I IFN production by human PDCs have been investigated in detail,14 the dynamic regulatory mechanism has not yet been clarified.
Both TLR7 and TLR9 are considered closely related with the high IFN-α level in systemic lupus erythematosus.2,3 However, earlier studies indicated that TLR7 activation, in contrast to TLR9 activation, led to sex-dependent IFN-α induction,15 highlighting the importance of TLR7 signaling for the development of the female-predominant autoimmune disease. So, we focused on the TLR7 pathway in PDCs.
Recently, microRNAs (miRNAs) have emerged as a major class of gene-expression regulators linked to most biological functions.16 They posttranscriptionally regulate gene expression by imperfectly base pairing with the 3′ untranslated regions (3′-UTRs) of target mRNAs, preventing protein accumulation by repressing their translation or accelerating mRNA degradation.16 miRNA biogenesis in animals has been extensively studied. Transcripts from genes encoding miRNAs, designated primary miRNAs (pri-miRNAs), are processed to precursor miRNA (pre-miRNA) hairpins, and then the pre-miRNAs are transported to the cytoplasm, where they are cleaved by Dicer, resulting in miRNA duplexes of approximately 21-23 nucleotides.16 The miRNA strands are selectively loaded into the RNA-induced silencing complex (RISC).16,17 For most miRNAs, the accumulation of the 2 strands is asymmetric. The more abundant strand is referred to as the miRNA, whereas its rarer partner is known as the star-form miRNA (miRNA*) species.17,18 Although miRNA has been studied extensively, the functions of miRNA* have so far been ignored because they are expressed relatively rarely.19 However, earlier research suggested that miRNA* species also have important functions in the miRNA regulatory network.18-20
Some miRNAs have been identified as important regulators of the immune system. Of these, miR-146a, miR-155, and miR-132, which are induced by TLR, are negative feedback regulators of the immune response.21-23 miR-146a has also been reported to negatively regulate type I IFN production in macrophages and peripheral blood mononuclear cells (PBMCs).24,25 This suggests that miRNAs are important players in TLR-induced type I IFN production. The questions raised are whether miRNAs play a role in human type I IFN production by PDCs, and if so, how miRNAs are involved in its dynamic regulation.
In this study, we used a Taqman Human MicroRNA Arrays–based screen to identify the miRNAs induced during primary human PDC activation, and found that miR-155 and its star-form partner, miR-155*, were most highly induced in the early stage of TLR7 stimulation. We also demonstrated that miR-155* positively regulates PDC IFN-α/β production by targeting IRAKM, whereas miR-155 plays a negative role by targeting TAB2. We have demonstrated, for the first time, how a miRNA cooperates with its star-form partner to dynamically regulate PDC IFN-α/β production by monitoring their kinetic changes together with those of their respective targets. We also found that 2 factors contribute to the opposite changes of miR-155* and miR-155 in the later stage of stimulation: First, IFN-α promoted miR-155 production, but inhibited miR-155* expression, and second, TLR7-activated KHSRP promoted miR-155 maturation, but reduced miR-155* maturation. The induction of KHSRP by autocrine/paracrine type I IFN further increased the difference in the accumulation of miR-155* and miR-155. Therefore, our finding suggests that the posttranscriptional regulation of a single pre-miRNA must be precisely regulated by external stimuli to adapt it to fulfill desired functions in the different stages of stimulation.
Methods
Isolation of PDCs
Fresh PBMCs were obtained from 200 healthy individuals. PDCs were isolated by magnetic-activated cell sorting using the Human Diamond Plasmacytoid Dendritic Cell Isolation kit (Miltenyi Biotec). The purity of the isolated PDCs was > 95%, as determined by flow cytometry. Ethics approval for this study was obtained from the Ethics Committee of Shanghai Renji Hospital, Shanghai Jiaotong University School of Medicine.
Cell culture, stimulation, and transfection
HeLa cells were kept under standard cell culture conditions in complete RPMI 1640 medium (Gibco, Invitrogen), containing 10% fetal calf serum, 100 U/mL penicillin (Gibco), and 100 μg/mL streptomycin (Gibco). Isolated PDCs were cultured in 96-well flat-bottomed plates at a concentration of 1 × 105 cells in 100 μL of complete RPMI 1640 medium. The PDCs were stimulated with 5 or 10 μg/mL R837. The final concentration of IFN-α used was 1000 U/mL. For pathway screening, PDCs were stimulated with 10 μg/mL R837 alone or in the presence of various inhibitors, including 2μM LY294002 (Tocris), 10μM pyrrolidine dithiocarbamate (Tocris), 25 μg/mL SP600125 (Tocris), or 10μM SB203580 (Tocris).
Small-interfering RNA (siRNA) and miRNA mimics or inhibitors were synthesized by Genepharma. PDCs were transfected with these RNA using Lipofectamine 2000 (Invitrogen). Unless otherwise indicated, 200 ng of nucleic acid complex was added together with 0.5 μL of Lipofectamine to the PDCs in a final volume of 150 μL. Six hours later, supernatants were removed and replaced with fresh complete RPMI 1640 medium containing 10 ng/mL recombinant human IL-3 (R&D Systems) for the next experiments.
TaqMan MicroRNA Arrays
Total RNA was isolated with the mirVana miRNA Isolation kit (Ambion). Samples for the TaqMan Human MicroRNA Arrays (Applied Biosystems) were reverse transcribed using Megaplex RT Primers and the TaqMan MicroRNA Reverse Transcription kit. The cDNAs were produced in preamplification reactions for 12 cycles using the Megaplex PreAmp Primers and TaqMan PreAmp Master Mix. The miRNA expression was then detected by real-time polymerase chain reactions (PCRs) with the TaqMan Universal PCR Master Mix (No AmpErase UNG) and TaqMan MicroRNA Array. The microarray data have been deposited in the Gene ExpressionOmnibus public database under accession number GSE21160.
Quantitative real-time RT-PCR
Total cellular RNA was isolated with TRIzol reagent (Invitrogen). cDNA was prepared by reverse transcription (PrimeScript RT Reagent kit; Takara) and amplified by real-time quantitative PCR (qPCR) with the primers shown in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Equal amounts of cDNA were used for the subsequent qPCR performed with the SYBR PrimeScript reverse-transcription (RT)–PCR kit (Takara). Amplification was performed in an ABI PRISM 7900 Real Time PCR System (Applied Biosystems). The amplification efficiency of these genes was the same as that for Rpl13a, as indicated by the standard curves for amplification, allowing us to use the following formula: fold difference = 2–(ΔCtA −ΔCtB), where Ct is the cycle threshold. miRNA expression was quantified using the TaqMan MicroRNA Expression Assay (Applied Biosystems), according to the manufacturer's protocol. miRNA expression was normalized to that of the endogenous RNU48.
Normalization and filtering
Sequence Detection Systems 2.3 software (Applied Biosystems) was used to analyze the qRT-PCR data. For the microarray data, miRNAs that were unreliably quantifiable or not expressed (Ct values of 30 or higher) in both the stimulated and control samples were excluded from further analysis, leaving a set of 174 miRNAs. RNU48 was used for internal normalization because it showed the most consistent expression in our cohort. The relative quantification method, 2–ΔCt, was used to calculate miRNA expression relative to that of RNU48. The fold induction for each miRNA was calculated using the following formula: fold induction = 2−(ΔCt stimulated PDC−ΔCt control PDC).
miRNA target prediction
Targetscan Version 5.1 (http://www.targetscan.org/vert_50, accessed October 2009) was used to predict the miR-155 targets. We used Targetscan Version 5.1 Custom (http://www.targetscan.org/vert_50/seedmatch.html, accessed October 2009) to predict the miR-155* targets. The details of the complete computational protocol are available at those sites and elsewhere.26
Luciferase assays
To test whether miR-155* would directly target IRAKM, HeLa cells plated in 96-well, flat-bottomed plates were transiently cotransfected with 50 ng of each reporter construct (wild-type and mutant IRAKM 3′-UTR and the psiCHECK-2 vector) and the synthetic miR-155* oligonucleotide or control oligonucleotide using Lipofectamine 2000 reagent (Invitrogen). Firefly and Renilla luciferase activities were determined 24 hours after transfection using the Dual-Luciferase Reporter Assay System (Promega). The values were normalized to firefly luciferase.
RNA immunoprecipitation assay
RNA immunoprecipitation (RIP) assays were performed as described by Chen et al27 and Ruggiero et al.28 Briefly, 1 × 106 primary PDCs were treated with R837 (10 μg/mL) for indicated times and were lysed by RIPA lysis and extraction buffer containing 10mM ribonucleoside-vanadyl complex (New England Biolabs). Lysates were spun at 14 000g for 15 minutes at 4°C, and supernatants were incubated overnight with antibodies at 4°C under rotation. Protein A/G–Sepharose (Yuekebio) was added for 4 hours at 4°C, and then the precipitates were washed 3× in lysis buffer and collected. RNA in the precipitates or supernatants was extracted with TRIzol reagent (Invitrogen). Equal amounts of RNA were reverse transcribed and subjected to PCR.
Results
Identification of miRNA induction during primary human PDC activation
To study whether miRNAs are involved in PDC activation, we detected miRNA changes in PDCs during TLR7 stimulation using Taqman Human MicroRNA Arrays. Considering that all the IFN-α subtype mRNAs reached peak levels 4 hours after TLR7 stimulation,29 we focused on the miRNAs that were induced in the early stage. Primary human PDCs purified from the PBMCs of 10 healthy donors were stimulated for 4 hours with R837, and the expression levels of mature miRNAs were measured. To ensure the reliability of the results, we first measured the mRNA expression in the same samples by qRT-PCR. Twelve IFN-α subtypes and IFN-β were successfully induced (supplemental Table 2). The cytokine TNF-α and chemokines (CCL3 and CCL4) were also successfully induced (supplemental Table 2). This is consistent with an earlier report.29
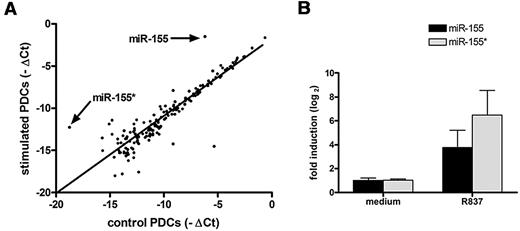
Microarray analysis of the miRNAs showed that 174 of the 657 miRNAs screened were expressed in the PDCs (supplemental Table 3). Among these 174 miRNAs, 5 miRNAs were significantly increased (more than 4-fold), whereas 14 were significantly reduced (more than 4-fold; supplemental Table 3). Furthermore, 4 of these 19 miRNAs with clearly altered levels of expression were star-form miRNAs. The existence of star-form miRNAs with altered expression indicates that they are also important regulators of the process. Compared with the other 17 miRNAs, miR-155* and miR-155 showed the highest induction (Figure 1A). qRT-PCR confirmed that miR-155 was induced 16-fold and miR-155* was induced 64-fold 4 hours after stimulation with TLR7 (Figure 1B). Because miR-155* and miR-155 were the most highly induced miRNAs, we considered that they might play an important role in PDC activation.
miR-155* and miR-155 show the highest induction in human PDCs upon TLR7 stimulation. (A) Taqman Human MicroRNA Arrays analysis of miRNA expression in PDCs after stimulation with R837. Human PDCs were stimulated with 10 μg/mL R837 for 4 hours. RNA was extracted and used for a microarray analysis to determine the expression levels of 657 human miRNAs. Data are presented on a scatter plot showing averaged log2-transformed relative expression (-▵Ct) performed on 2 different PDC preparations of each miRNA for media control (x-axis) and sample stimulated with R837 (y-axis). (B) The RNA used in panel A was analyzed by TaqMan MicroRNA Expression Assay to assay the expression of miR-155* and miR-155. Expression levels were normalized to RNU48.
miR-155* and miR-155 show the highest induction in human PDCs upon TLR7 stimulation. (A) Taqman Human MicroRNA Arrays analysis of miRNA expression in PDCs after stimulation with R837. Human PDCs were stimulated with 10 μg/mL R837 for 4 hours. RNA was extracted and used for a microarray analysis to determine the expression levels of 657 human miRNAs. Data are presented on a scatter plot showing averaged log2-transformed relative expression (-▵Ct) performed on 2 different PDC preparations of each miRNA for media control (x-axis) and sample stimulated with R837 (y-axis). (B) The RNA used in panel A was analyzed by TaqMan MicroRNA Expression Assay to assay the expression of miR-155* and miR-155. Expression levels were normalized to RNU48.
miR-155* and miR-155 are induced by TLR7 through the c-Jun N-terminal kinase (JNK) pathway
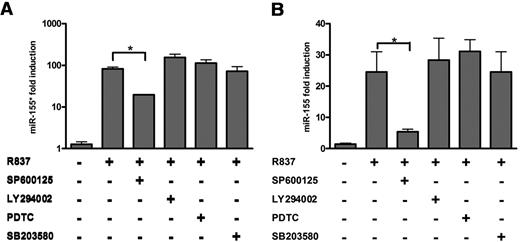
It has been demonstrated that TLR stimulation can lead to the activation of the MyD88/TRAF6/IRAK1/4 pathway, after which the PI3K-AKT, p38, JNK-AP1 (activator protein 1), and nuclear factor (NF)–κB pathways are activated.30 In a previous study, the promoter region of pri-miR-155 was reported to contain putative NF-κB– and AP1-binding sites.31 In addition to the NF-κB pathway, both the PI3K and JNK pathways also induce pri-miR-155 production.31-33 To identify the pathways that regulate miR-155* and miR-155 expression in PDCs, we pretreated primary PDCs with an NF-κB inhibitor (PDTC), a PI3K inhibitor (LY294002), a p38 inhibitor (SB203580), or a JNK inhibitor (SP600125) and then exposed them to R837 for 8 hours. The results showed that only JNK inhibition reduced the induction of miR-155* and miR-155 (Figure 2A-B). Therefore, we concluded that TLR7 induces miR-155* and miR-155 expression through the JNK pathway in primary human PDCs.
miR-155* and miR-155 are induced by TLR7 in PDCs via the JNK-signaling pathway. PDCs were cultured for 8 hours with medium or R837 in the presence or absence of LY294002, PDTC, SP600125, or SB203580. miR-155* (A) and miR-155 (B) were analyzed by TaqMan MicroRNA Expression Assay and normalized to RNU48 levels. The data are representative of at least 3 independent experiments, each based on a different PDC preparation. The data were analyzed with 2-tailed Student t test. *P < .05.
miR-155* and miR-155 are induced by TLR7 in PDCs via the JNK-signaling pathway. PDCs were cultured for 8 hours with medium or R837 in the presence or absence of LY294002, PDTC, SP600125, or SB203580. miR-155* (A) and miR-155 (B) were analyzed by TaqMan MicroRNA Expression Assay and normalized to RNU48 levels. The data are representative of at least 3 independent experiments, each based on a different PDC preparation. The data were analyzed with 2-tailed Student t test. *P < .05.
miR-155* and miR-155 have opposite effects on TLR7-mediated type I IFN production in human PDCs
miR-155 is a key regulator of innate immunity,34 but its specific function in activated human PDCs remains unclear. miR-155*, which is cleaved from the same precursor as miR-155, has never been studied before. We devised a strategy to overexpress miR-155* and miR-155 by introducing 2′-O-methyl–modified double-stranded RNA for the specific generation of mature miR-155* or miR-155 (miR-155* mimic or miR-155 mimic, respectively) in PDCs. To functionally inhibit miR-155* and miR-155, 2′-O-methyl–modified oligonucleotides specifically designed to knock down miR-155* or miR-155 expression (as-miR-155* or as-miR-155, respectively) were used.
We evaluated the overexpression of miR-155* and miR-155 after the transfection of their respective mimics into primary PDCs. Both miR-155* and miR-155 were overexpressed approximately 100-fold 24 hours after transfection, when their expression was analyzed with the TaqMan MicroRNA Expression Assay (supplemental Figure 1A-B). The efficiency and specificity of miR-155* suppression by as-miR-155* and miR-155 suppression by as-miR-155 were also tested by introducing them into primary human PDCs. Reductions of approximately 30-fold in the miR-155 and miR-155* levels were achieved with their respective inhibitors after activation with R837. However, as-miR-155 had no effect on miR-155* expression, and as-miR-155* had no effect on miR-155 expression (supplemental Figure 1C-D).
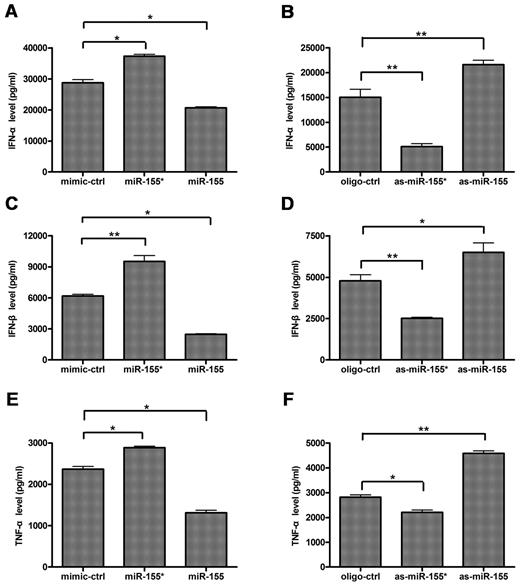
To investigate the roles of miR-155* and miR-155 in type I IFN production by PDCs stimulated with TLR7, we transfected purified primary human PDCs with miR-155* mimic, as-miR-155*, miR-155 mimic, or as-miR-155 before stimulation with R837. IFN-α and IFN-β expression were measured and compared with those of cells transfected with the mimic control or the oligonucleotide control. The introduction of miR-155* mimic augmented IFN-α/β expression, whereas as-miR-155* suppressed their expression (Figure 3A-D). In contrast, the introduction of miR-155 mimic suppressed IFN-α/β expression, whereas as-miR-155 augmented them. It should be highlighted that as-miR-155* inhibited up to 70% of IFN-α production (Figure 3B). These results suggest that both miR-155* and miR-155 are important in the regulation of type I IFN production by PDCs. The activation of PDCs by TLR7 induced the expression of the proinflammatory cytokine, TNF-α, in addition to large amounts of type I IFN.1 Like their opposite effects on IFN-α/β production, miR-155* augmented TNF-α expression, whereas miR-155 suppressed it in TLR7-activated PDCs (Figure 3E-F). Taking these data together, we conclude that miR-155* and miR-155 exert opposite effects on TLR7-mediated type I IFN production in primary human PDCs.
miR-155* and miR-155 have opposite effects on TLR7-mediated IFN-α/β and TNF-α production in human PDCs. Human PDCs were transfected with (A,C,E) mimic control (mimic-ctrl), miR-155 mimic (miR-155), or miR-155* mimic (miR-155*), (B,D,F) oligonucleotide control (oligo-ctrl), as-miR-155, or as-miR-155*, with Lipofectamine 2000. Eight hours after transfection, the PDCs were stimulated with 10 μg/mL R837 for 24 hours. IFN-α (A-B), IFNβ (C-D), and TNF-α (E-F) expression levels in the supernatants were measured by enzyme-linked immunosorbent assay (ELISA). The data are mean ± SD of triplicate wells from 1 of 3 experiments with similar results, analyzed with 2-tailed Student t test. *P < .05; **P < .01.
miR-155* and miR-155 have opposite effects on TLR7-mediated IFN-α/β and TNF-α production in human PDCs. Human PDCs were transfected with (A,C,E) mimic control (mimic-ctrl), miR-155 mimic (miR-155), or miR-155* mimic (miR-155*), (B,D,F) oligonucleotide control (oligo-ctrl), as-miR-155, or as-miR-155*, with Lipofectamine 2000. Eight hours after transfection, the PDCs were stimulated with 10 μg/mL R837 for 24 hours. IFN-α (A-B), IFNβ (C-D), and TNF-α (E-F) expression levels in the supernatants were measured by enzyme-linked immunosorbent assay (ELISA). The data are mean ± SD of triplicate wells from 1 of 3 experiments with similar results, analyzed with 2-tailed Student t test. *P < .05; **P < .01.
miR-155* targets IRAKM and miR-155 targets TAB2
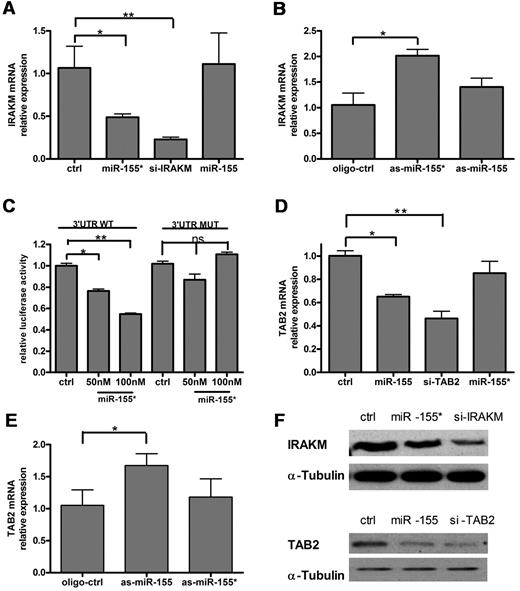
To identify the potential targets of miR-155*, we used the miRNA target identification program, Targetscan Version 5.1.26 This analysis identified IRAKM as a potential target for miR-155* (supplemental Figure 2A). To confirm that IRAKM is regulated by miR-155*, we transfected primary human PDCs with miR-155* mimic or with as-miR-155*. qRT-PCR revealed that miR-155* inhibited the expression of IRAKM mRNA (Figure 4A), whereas as-miR-155* enhanced it (Figure 4B). Further study demonstrated that miR-155* inhibited IRAKM protein expression (Figure 4F). To confirm the prediction that miR-155* interacts with the 3′-UTR of IRAKM, we cloned 1000 bp of the 3′-UTR of IRAKM, which included the miR-155*–binding site, downstream of the Renilla luciferase gene, and cotransfected it with various concentrations of miR-155* into HeLa cells. The results indicated that miR-155* suppressed Renilla luciferase activity in a dose-dependent manner, and that the mutation in the seed sequence (supplemental Figure 2A) of the IRAKM 3′-UTR, where miR-155* is predicted to bind, abolished the suppression of Renilla luciferase activity by miR-155* (Figure 4C).
miR-155* targets IRAKM and miR-155 targets TAB2. Human PDCs were transfected with (A,D) mimic control (ctrl), miR-155 mimic (miR-155), miR-155* mimic (miR-155*), IRAKM siRNA (si-IRAKM), or TAB2 siRNA (si-TAB2), (B,E) oligonucleotide control (oligo-ctrl), as-miR-155, or as-miR-155*, and then stimulated with R837 for 16 hours. qRT-PCR analysis of IRAKM and TAB2 mRNA expression was normalized to Rpl13a RNA levels. These graphs show the relative expression of IRAKM mRNA (A-B) and TAB2 mRNA (D-E), normalized to the control level. The data are the means (± SD) of triplicate qRT-PCR, representing at least 3 independent experiments, each based on a different PDC preparation. (C) miR-155 directly suppresses IRAKM mRNA through 3′-UTR interactions. The 3′-UTR of human IRAKM was cloned into the psiCHECK-2 vector, downstream of the Renilla luciferase gene. The construct was then cotransfected into HeLa cells with the indicated amount of the miR-155* mimic (miR-155*) or mimic control (ctrl), and the luciferase activity was quantified. The graph shows the relative luciferase activity normalized to the mimic control values. The data are representative of at least 3 independent experiments. (D) miR-155* inhibits IRAKM protein expression, and miR-155 inhibits TAB2 protein expression. (Top) Western blot analysis of IRAKM protein expression in THP-1 cells 48 hours after transfection with IRAKM siRNA (si-IRAKM), miR-155* mimic (miR-155*), or mimic control (ctrl). (Bottom) Western blot analysis of IRAKM protein expression in THP-1 cells 48 hours after transfection with TAB2 siRNA (si-TAB2), miR-155 mimic (miR-155), or mimic control (ctrl). α-tubulin is shown to confirm equal loading. The data are representative of at least 3 independent experiments, each based on a different PDC preparation.
miR-155* targets IRAKM and miR-155 targets TAB2. Human PDCs were transfected with (A,D) mimic control (ctrl), miR-155 mimic (miR-155), miR-155* mimic (miR-155*), IRAKM siRNA (si-IRAKM), or TAB2 siRNA (si-TAB2), (B,E) oligonucleotide control (oligo-ctrl), as-miR-155, or as-miR-155*, and then stimulated with R837 for 16 hours. qRT-PCR analysis of IRAKM and TAB2 mRNA expression was normalized to Rpl13a RNA levels. These graphs show the relative expression of IRAKM mRNA (A-B) and TAB2 mRNA (D-E), normalized to the control level. The data are the means (± SD) of triplicate qRT-PCR, representing at least 3 independent experiments, each based on a different PDC preparation. (C) miR-155 directly suppresses IRAKM mRNA through 3′-UTR interactions. The 3′-UTR of human IRAKM was cloned into the psiCHECK-2 vector, downstream of the Renilla luciferase gene. The construct was then cotransfected into HeLa cells with the indicated amount of the miR-155* mimic (miR-155*) or mimic control (ctrl), and the luciferase activity was quantified. The graph shows the relative luciferase activity normalized to the mimic control values. The data are representative of at least 3 independent experiments. (D) miR-155* inhibits IRAKM protein expression, and miR-155 inhibits TAB2 protein expression. (Top) Western blot analysis of IRAKM protein expression in THP-1 cells 48 hours after transfection with IRAKM siRNA (si-IRAKM), miR-155* mimic (miR-155*), or mimic control (ctrl). (Bottom) Western blot analysis of IRAKM protein expression in THP-1 cells 48 hours after transfection with TAB2 siRNA (si-TAB2), miR-155 mimic (miR-155), or mimic control (ctrl). α-tubulin is shown to confirm equal loading. The data are representative of at least 3 independent experiments, each based on a different PDC preparation.
In an analysis using Targetscan Version 5.1,26 we identified TAB2 as a potential target for miR-155 (supplemental Figure 2B). It has been demonstrated that TAB2 is suppressed directly by miR-155 in monocyte-derived dendritic cells.22 To confirm that miR-155 also regulates TAB2 in PDCs, we transfected human PDCs with miR-155 mimic or with as-miR-155. qRT-PCR showed that miR-155 inhibited the expression of TAB2 mRNA (Figure 4D), whereas as-miR-155 enhanced it (Figure 4E). Further study demonstrated that miR-155 inhibited TAB2 protein expression (Figure 4F). The results described in this section demonstrate that IRAKM and TAB2 are the direct targets of miR-155* and miR-155, respectively.
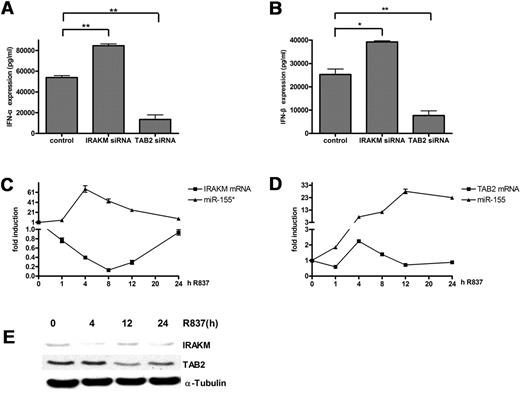
IRAKM negatively regulates the TLR pathways by preventing the dissociation of IRAK1 and IRAK4 from MyD88 and the formation of IRAK1/TRAF6 complexes.35 Although IRAKM has been reported to negatively regulate the TLR7 pathway,36 its function in PDCs during TLR7 stimulation has not yet been investigated. We demonstrated its negative role by transfecting a siRNA directed against IRAKM or a control siRNA into human PDCs. The results showed that the knockdown of IRAKM expression promoted IFN-α and IFN-β production (Figure 5A-B). TAB2, which was first identified as an adaptor linking transforming growth factor-α–activated kinase to TRAF6, has been reported to facilitate TRAF6 ubiquitination and IKK-α activation.37 Both TRAF6 and IKK-α play key roles in type I IFN production in PDCs upon TLR stimulation,38,39 so TAB2 may also regulate type I IFN production by PDCs. We demonstrated the positive role of TAB2 by transfecting TAB2 siRNA or control siRNA into human PDCs. Knockdown of TAB2 expression reduced IFN-α/β production (Figure 5A-B).
IRAKM and TAB2 negatively correlate with miR-155* and miR-155, respectively. (A-B) IRAKM and TAB2 had opposite effects on IFN-α and IFN-β production in human PDCs upon TLR7 stimulation. Purified human PDCs transfected with siRNA control, IRAKM siRNA, or TAB2 siRNA were stimulated with R837 for 24 hours. IFN-α (A) and IFN-β (B) in the supernatants were measured by ELISA. The data were from 1 of 3 experiments with similar results, analyzed with 2-tailed Student t test. *P < .05, **P < .01. (C-D) miR-155* and miR-155 correlated negatively with their respective targets during TLR7 stimulation. Purified PDCs were stimulated with 5 μg/mL R837 over a 24-hour time course and harvested at the indicated time points. miR-155* (C) and miR-155 (D) were analyzed with the TaqMan MicroRNA Expression Assay and normalized to the RNU48 levels. IRAKM (C) and TAB2 (D) mRNA were detected by qRT-PCR and normalized to the RPL13a RNA levels. These graphs show the fold induction calculated by normalizing the expression values at different time points to the 0-hour values. The data are representative of at least 3 independent experiments, each based on a different PDC preparation. (E) Immunoblot kinetics analysis for IRAKM and TAB2. An α-tubulin immunoblot is shown for equal loading control. Data are representative of 2 independent experiments.
IRAKM and TAB2 negatively correlate with miR-155* and miR-155, respectively. (A-B) IRAKM and TAB2 had opposite effects on IFN-α and IFN-β production in human PDCs upon TLR7 stimulation. Purified human PDCs transfected with siRNA control, IRAKM siRNA, or TAB2 siRNA were stimulated with R837 for 24 hours. IFN-α (A) and IFN-β (B) in the supernatants were measured by ELISA. The data were from 1 of 3 experiments with similar results, analyzed with 2-tailed Student t test. *P < .05, **P < .01. (C-D) miR-155* and miR-155 correlated negatively with their respective targets during TLR7 stimulation. Purified PDCs were stimulated with 5 μg/mL R837 over a 24-hour time course and harvested at the indicated time points. miR-155* (C) and miR-155 (D) were analyzed with the TaqMan MicroRNA Expression Assay and normalized to the RNU48 levels. IRAKM (C) and TAB2 (D) mRNA were detected by qRT-PCR and normalized to the RPL13a RNA levels. These graphs show the fold induction calculated by normalizing the expression values at different time points to the 0-hour values. The data are representative of at least 3 independent experiments, each based on a different PDC preparation. (E) Immunoblot kinetics analysis for IRAKM and TAB2. An α-tubulin immunoblot is shown for equal loading control. Data are representative of 2 independent experiments.
IRAKM negatively correlates with miR-155*, and TAB2 negatively correlates with miR-155
To study the correlation between the miRNAs and their targets during TLR7 stimulation, the dynamic changes of IRAKM, TAB2, mature miR-155*, and mature miR-155 were investigated. IRAKM mRNAs decreased continuously until the 8-hour time point and then gradually recovered to the starting level, whereas miR-155* increased rapidly, peaked at the 4-hour time point, and then decreased dramatically (Figure 5C). Similar to its mRNA changes, the protein levels of IRAKM decreased 4 hours poststimulation and recovered at the 12-hour time point (Figure 5E). As expected from the kinetics of miR-155, the protein level of TAB2 did not increase (Figure 5E) when its mRNA slightly increased at the 4-hour time point (Figure 5D). Then, both mRNA and protein levels of TAB2 decreased when miR-155 continued to increase significantly. These results suggest that miR-155* and miR-155 function through IRAKM and TAB2, respectively, in PDCs during TLR7 stimulation.
Different kinetics of miR-155* and miR-155 induced by TLR7 and IFN-α allow them to fine tune type I IFN production
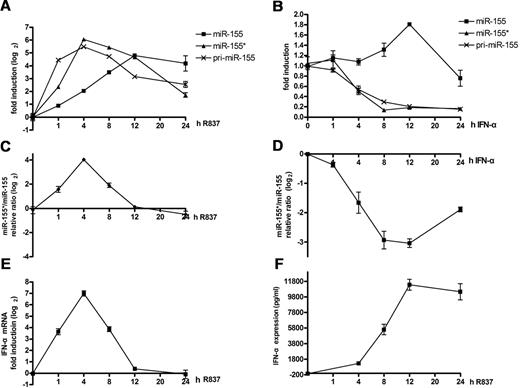
Because miR-155* and miR-155 have opposite effects, it is important to examine how they collaborate with each other in regulating IFN-α production by PDCs during TLR7 stimulation. Therefore, we monitored the kinetics of both of them when induced by TLR7 over a 24-hour time course. miR-155*, which showed a similar expression pattern to that of pri-miR-155, was rapidly induced, reached its peak level of approximately 60-fold induction at the 4-hour time point, and then decreased. However, miR-155 increased much more slowly, showing an induction of only 7-fold when miR-155* peaked. Then, miR-155 continued to increase and reached its peak level at the 12-hour time point, after which this high-level expression was maintained (Figure 6A). Considering that they were processed from a single precursor, the changes in the ratio of miR-155* to miR-155 might better reflect their combined effects. Our results show that their ratio increased rapidly, peaked at the 4-hour time point, and then declined rapidly (Figure 6C). As expected from the kinetics of this ratio, IFN-α mRNA was strongly induced, had increased more than 100-fold at the 4-hour time point, and then declined rapidly to the level observed before stimulation at the 12-hour time point (Figure 6E). From the results discussed in this paragraph, we infer that the more substantial increase in miR-155* expression in the early stage (before 4 hours) of PDC activation is part of the positive feedback that facilitates type I IFN production, whereas the rapid decline in miR-155* and the increase in miR-155 at a relatively later stage form part of the negative feedback regulation that circumvents excessive type I IFN production.
Different dynamic induction of miR-155* and miR-155 by TLR7 and IFNα. (A-B) Kinetic analysis of TLR7 and IFN-α induction of pri-miR-155, mature miR-155, and mature miR-155*. Human PDCs were stimulated with R837 (A) or IFN-α (B) over a 24-hour time course and harvested at the indicated time points. miR-155* and miR-155 were analyzed with the TaqMan MicroRNA Expression Assay and normalized to the RNU48 levels. Pri-miR-155 mRNA was detected by qRT-PCR and normalized to the RPL13a RNA levels. The graph shows the fold induction calculated by normalizing the relative expression values at different time points to the 0-hour values. (C-D) IFN-α and TLR ligands had opposite effects on the change in the ratios of miR-155* to miR-155. Purified PDCs were stimulated with either IFN-α or R837 over a 24-hour time course and harvested at the indicated time points. miR-155* and miR-155 were analyzed with the TaqMan MicroRNA Expression Assay and normalized to the RNU48 levels. The ratios of miR-155* to miR-155 were calculated. The graph shows the fold changes after the ratios at different time points were normalized to the 0-hour ratio. (E) IFN-α mRNAs from panel A were detected by qRT-PCR and normalized to RPL13a RNA levels. (F) IFN-α in the supernatant of panel A was detected by ELISA. The data are representative of at least 3 independent experiments, each based on a different PDC preparation.
Different dynamic induction of miR-155* and miR-155 by TLR7 and IFNα. (A-B) Kinetic analysis of TLR7 and IFN-α induction of pri-miR-155, mature miR-155, and mature miR-155*. Human PDCs were stimulated with R837 (A) or IFN-α (B) over a 24-hour time course and harvested at the indicated time points. miR-155* and miR-155 were analyzed with the TaqMan MicroRNA Expression Assay and normalized to the RNU48 levels. Pri-miR-155 mRNA was detected by qRT-PCR and normalized to the RPL13a RNA levels. The graph shows the fold induction calculated by normalizing the relative expression values at different time points to the 0-hour values. (C-D) IFN-α and TLR ligands had opposite effects on the change in the ratios of miR-155* to miR-155. Purified PDCs were stimulated with either IFN-α or R837 over a 24-hour time course and harvested at the indicated time points. miR-155* and miR-155 were analyzed with the TaqMan MicroRNA Expression Assay and normalized to the RNU48 levels. The ratios of miR-155* to miR-155 were calculated. The graph shows the fold changes after the ratios at different time points were normalized to the 0-hour ratio. (E) IFN-α mRNAs from panel A were detected by qRT-PCR and normalized to RPL13a RNA levels. (F) IFN-α in the supernatant of panel A was detected by ELISA. The data are representative of at least 3 independent experiments, each based on a different PDC preparation.
Type I IFN has been reported to facilitate TLR7/9-induced PDC activation.40,41 Considering that IFN-α was detected in the supernatant as early as 4 hours after TLR7 stimulation (Figure 6F), we next examined whether type I IFN is involved in the regulation of miR-155* and miR-155 expression. An earlier study demonstrated that IFN-α can induce the expression of pri-miR-155 and miR-155 indirectly through TNF-α in macrophages,33 so we tested whether it would play the same role in human PDCs and if miR-155* would be regulated in the same way. To our surprise, the expression of miR-155 was slightly up-regulated (less than 1-fold), whereas that of miR-155* and pri-miR-155 were down-regulated by stimulation with IFN-α over a 24-hour time course (Figure 6B). These kinetics were somewhat similar to those in the later stage of TLR7 stimulation, between the 4- and 12-hour time points. Moreover, because TNF-α was not detected in the supernatant until 12 hours after stimulation by IFN-α (supplemental Figure 3A), and blocking TNF-α did not eliminate the IFN-α–induced increase of miR-155 (supplemental Figure 3B) and decrease of miR-155* (supplemental Figure 3C), it is unlikely that the induction of miR-155 by IFN-α is attributable to the autocrine/paracrine TNF-α. Further analysis indicated that the ratio of miR-155* to miR-155 changed in opposite directions when stimulated with TLR7, compared with IFN-α (Figure 6D).
To confirm that the autocrine/paracrine type I IFN is involved in the regulation of miR-155* and miR-155 during TLR7 stimulation, we stimulated PDCs with R837 alone or in the presence of blocking receptor for type I IFN (anti-IFN). The ratio between miR-155* and miR-155 was calculated and increased when the type I IFN functions were blocked during TLR7 stimulation (supplemental Figure 3D). This indicates that type I IFN derived from TLR7-stimulated PDCs might regulate their own production through its impact on miR-155* and miR-155 expression.
KHSRP induces opposite kinetics of miR-155* and miR-155 expression in the later stage of TLR7 stimulation
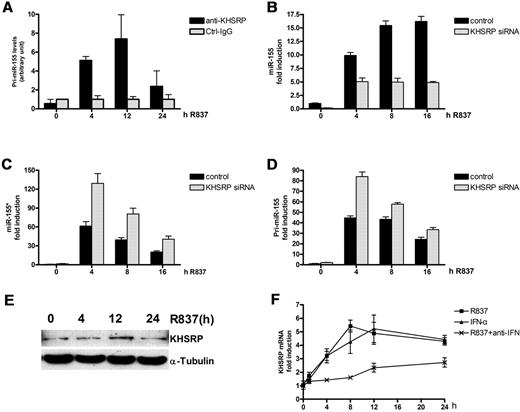
Although autocrine/paracrine type I IFN is involved in the different regulation of miR-155* and miR-155, blocking of type I IFN function could not completely eliminate their different changes (supplemental Figure 3D). So, other mechanisms may also participate in the process. miRNAs can be regulated at the transcriptional and posttranscriptional levels.16,42 Because both miR-155* and miR-155 are processed from pre-miR-155, it is more likely that their different changes in the later stage of TLR7 stimulation resulted from posttranscriptional regulation. KHSRP binds to the terminal loop of the miR-155 precursor and promotes its maturation upon TLR stimulation in macrophage.28 However, its effect on miR-155* expression has not been studied. First, RIP assays were performed to evaluate the direct interaction of KHSRP with pri-miR-155 in human primary PDCs. Results showed that KHSRP binds to pri-miR-155 in a time-dependent manner (Figure 7A). To determine whether KHSRP participates in the regulation of miR-155* and miR-155 in PDCs, we transfected KHSRP siRNA into primary human PDCs. The efficiency of the knockdown of KHSRP mRNA and protein by siRNA was confirmed (supplemental Figure 4A-B). A kinetic analysis demonstrated that the knockdown of KHSRP greatly impaired the expression of mature miR-155 (Figure 7B), whereas it increased the expression of miR-155* (Figure 7C) and pri-miR-155 (Figure 7D). This indicates that KHSRP leads to opposite dynamic changes in miR-155* and miR-155 in the later stage of PDC stimulation by TLR7.
KHSRP promotes miR-155 maturation, but inhibits miR-155* production. (A) Anti-KHSRP antibody immunoprecipitates pri-miR-155. Primary human PDCs were treated with R837 for indicated times and lysed, and total cell extracts were immunoprecipitated, as indicated. Control immunoglobulin G (IgG; Ctrl-IgG) was used as the negative control. RNA was purified from immunocomplexes and analyzed by qRT-PCR. (B-D) Human PDCs transfected with either the control siRNA or KHSRP siRNA were stimulated with R837 over a 16-hour time course and harvested at the indicated time points. miR-155 (B) and miR-155* (C) were analyzed with the TaqMan MicroRNA Expression Assay and normalized to the RNU48 levels. Pri-miR-155 mRNA (D) was detected by qRT-PCR and normalized to the RPL13a RNA levels. These graphs show the fold induction calculated by normalizing the expression values at different time points to the control 0-hour values. (E) Immunoblot kinetics analysis for KHSRP. An α-tubulin immunoblot is shown for equal loading control. Data are representative of 2 independent experiments. (F) KHSRP was induced by R837 and IFN-α. Kinetic analysis of the induction of KHSRP mRNA by R837 and IFN-α. Human PDCs were stimulated with IFN-α or R837 alone or in the presence of 5 μg/mL type I IFN blocking receptor B18R (anti-IFN) over a 24-hour time course and harvested at the indicated time points. KHSRP mRNA was detected by qRT-PCR and normalized to RPL13a RNA levels. The graph shows the fold induction calculated by normalizing the expression values at different time points to the 0-hour values. The data are representative of at least 3 independent experiments, each based on a different PDC preparation.
KHSRP promotes miR-155 maturation, but inhibits miR-155* production. (A) Anti-KHSRP antibody immunoprecipitates pri-miR-155. Primary human PDCs were treated with R837 for indicated times and lysed, and total cell extracts were immunoprecipitated, as indicated. Control immunoglobulin G (IgG; Ctrl-IgG) was used as the negative control. RNA was purified from immunocomplexes and analyzed by qRT-PCR. (B-D) Human PDCs transfected with either the control siRNA or KHSRP siRNA were stimulated with R837 over a 16-hour time course and harvested at the indicated time points. miR-155 (B) and miR-155* (C) were analyzed with the TaqMan MicroRNA Expression Assay and normalized to the RNU48 levels. Pri-miR-155 mRNA (D) was detected by qRT-PCR and normalized to the RPL13a RNA levels. These graphs show the fold induction calculated by normalizing the expression values at different time points to the control 0-hour values. (E) Immunoblot kinetics analysis for KHSRP. An α-tubulin immunoblot is shown for equal loading control. Data are representative of 2 independent experiments. (F) KHSRP was induced by R837 and IFN-α. Kinetic analysis of the induction of KHSRP mRNA by R837 and IFN-α. Human PDCs were stimulated with IFN-α or R837 alone or in the presence of 5 μg/mL type I IFN blocking receptor B18R (anti-IFN) over a 24-hour time course and harvested at the indicated time points. KHSRP mRNA was detected by qRT-PCR and normalized to RPL13a RNA levels. The graph shows the fold induction calculated by normalizing the expression values at different time points to the 0-hour values. The data are representative of at least 3 independent experiments, each based on a different PDC preparation.
We also found that KHSRP is induced by TLR7 and IFN-α (Figure 7E-F). Reduced levels of KHSRP mRNA were detected when the autocrine/paracrine type I IFN functions were blocked. This suggests that type I IFN enhances the opposite changes in miR-155* and miR-155 expression through its impact on KHSRP expression.
We propose an overall model for the functions of miR-155* and miR-155 in PDCs (supplemental Figure 5). In the initial stage, both miR-155* and miR-155 are induced by the TLR7–JNK pathway. miR-155*, which targets IRAKM, increases more rapidly than miR-155, facilitating IFN-α/β production. In the later stage, under the influence of TLR7-derived type I IFN and TLR7-activated KHSRP, miR-155 increases quickly, whereas miR-155* decreases sharply. Moreover, type I IFN further expands the difference in the levels of miR-155 and miR-155* by the induction of KHSRP expression. miR-155 negatively regulates IFN-α/β production by repressing TAB2 and protects PDCs from excessive response to TLR stimulation.
Discussion
The activation of PDCs by TLR7/9 is associated with changes in the expression of positive and negative regulators.2 Although much progress has been made toward a better understanding of the molecular elements that act in this innate activation pathway and support the critical functions of PDCs, further comprehensive and sensitive analyses at the molecular level in new fields should provide new insights in the regulation of PDC function.6 The roles of miRNAs, which have a broad, important role in the immune system,43 have not previously been studied in PDCs. One finding of this study is that miR-155* and miR-155 cooperate to dynamically fine tune type I IFN production. Earlier studies have found that pairs of molecules from a single precursor have opposite functions in the same pathway, such as MyD88/MyD88s, TLR2/sTLR2, and IRAK2/IRAK2cd.44 Our results are consistent with the idea that alternative splicing plays an important role in the fine tuning of TLR responses.
Our finding that miR-155* and miR-155 are the most strongly induced miRNAs in primary human PDCs led us to speculate that they might be important in PDC activation. Further investigation demonstrated that they have opposite effects in regulating type I IFN production in PDCs. Consistent with their functions, they are also differentially induced by TLR7 stimulation. miR-155* is mainly induced in the early stage, whereas miR-155 peaks in the later stage post-TLR7 stimulation. Because IFN-α mRNA decreased rapidly 4 hours after stimulation, it is apparent that the positive regulation upstream of IFN-α gradually weakens and the negative regulation becomes dominant after the 4-hour time point. The fully consistent changes in mature miR-155* and IFN-α mRNA expression provide further strong evidence for the positive effect of miR-155* on IFN-α production. However, the continuous increase in miR-155, together with the reduction in TAB2 and the recovery of IRAKM, provide a good explanation of the rapid reduction in IFN-α mRNA levels. It has been reported recently that miR-155 directly suppresses MyD88 protein expression.45 Because MyD88 is an important component for TLR7/9-induced PDC activation,6,38 our findings are further supported by this report.
Besides miR-155, miR-146a, an important negative regulator of the TLR pathway,21 could also have been induced by R837 in primary PDCs (supplemental Figure 6), albeit to a lesser extent. It implies that miR-146a may also participate in regulating PDC activation.
It has been widely assumed that the miRNA* species simply promotes the accurate processing of its miRNA partner, and that miRNAs*, because of a certain degree of imprecision in miRNA strand selection, might be neutral and thus tolerated in vivo.17,46 However, other reports have suggested that miRNA* species are evolutionarily conserved and have inhibitory activity like that of miRNA in both cultured cells and transgenic animals.19,20 Here, we have identified miR-155* as an important player in PDC type I IFN production. More importantly, we have demonstrated, for the first time, how an miRNA* cooperates with its miRNA partner in the same physiological process. In addition to PDCs, miR-155* could also have been induced by lipopolysaccharide and copolymer of polyinosinic and polycytidylic acids in monocyte-derived dendritic cells (moDCs) in a similar way (supplemental Figure 7A-B). Also, inhibition of miR-155* in moDCs decreased the lipopolysaccharide-induced expression of TNF-α (supplemental Figure 7C). Our findings not only extend the known miRNA regulatory network in the immune systems, but emphasize the importance of the cooperation between miRNA* and miRNA.
The posttranscriptional regulation of miRNA has been extensively studied.16,40 For most miRNAs, only 1 guide strand is loaded into the RNA-induced silencing complex, whereas the other strand is rapidly destroyed.16,17 However, 31 of the 174 detected miRNAs were star-form miRNAs, and among these, 22 pairs of miRNA and miRNA* were both detected in primary human PDCs (supplemental Table 3). This indicates that both strands transcribed from the pre-miRNA are selected, in some cases. Furthermore, 9 miRNAs* were detected in 2 independent PDC preparations, whereas their nonstar-form partners were not (supplemental Table 3). These results suggest that factors other than the stability of both termini of the dsRNA also affect the strand selection of a pre-miRNA. Here, we have shown that miR-155* and miR-155 are differentially regulated by KHSRP and type I IFN. The type I IFN also causes more KHSRP activation by TLR7 through its induction of KHSRP expression, which further expands the difference between miR-155* and miR-155 expression in the later stage of PDC activation. We thus show, for the first time, that both strands of a pre-miRNA can be strictly regulated by external stimulus, allowing them to dynamically fine tune the host response. We propose that the strand selection of a pre-miRNA may be another important posttranscriptional regulatory mechanism, which diversifies the functional incorporation of the miRNAs into endogenous regulatory networks.
Host-derived self-RNA released from damaged cells normally fails to activate PDCs.2 However, self-RNA molecules, particularly those rich in uridine or uridine and guanosine and those in small nuclear ribonucleoproteins, can trigger PDCs to produce type I IFN via TLR7 when delivered to endosomes by autoantibodies or liposomes.2,47 The development of autoimmune disease, such as systemic lupus erythematosus is associated with enhanced PDC activation and type I IFN production.2,3,6 Understanding the functions of miR-155* and miR-155 in PDC activation should contribute to a better understanding of immune homeostasis and to the identification of novel therapeutic targets for the treatment of autoimmune disorders.
As well as in inflammatory response, PDCs have been suggested to play immunosuppressive roles in cancer tissues that have an impaired capacity to produce IFN-α in tumor immunity.5,48 miR-155 is consistently up-regulated in tumor cells.34,49,50 Therefore, the inhibition of miR-155 should not only promote PDC type I IFN production, breaking tolerance in tumors, but also prevent the proliferation of the tumor cells themselves.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported, in part, by the National High Technology Research and Development Program of China (863 Program; no. 2007AA02Z123), the National Basic Research Program of China (973 Program; no. 2007CB947900), the National Natural Science Foundation of China (Nos. 30700734, 30301026, 30971632, and 30801026), the Program of the Shanghai Commission of Science and Technology (nos. 06JC14050, 07ZR14130, 08JC1414700, and 10JC1409300), the Doctoral Innovation Fund of Shanghai Jiao Tong University School of Medicine (BXJ201020), and the Program of Shanghai Subject Chief Scientist (no. 07XD14021). L.W. was supported by a fellowship and Project Grants from the Australian National Health and Medical Research Council.
Authorship
Contribution: N.S., H.Z., X.H., and S.C. contributed to study design; H.Z., X.H., H.C., and X.L. contributed to acquisition of data; H.Z., X.H., H.C., X.L., Y.T., N.S., and L.W. contributed to analysis and interpretation of data; H.Z., N.S., L.W., and X.H. contributed to manuscript preparation; and H.Z. and X.H. performed statistical analysis. N.S. had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nan Shen, Department of Rheumatology, Renji Hospital, Shanghai Jiaotong University School of Medicine, 145 Shan Dong Rd (Central), Shanghai 200001, China; e-mail: nanshensibs@gmail.com.
References
Author notes
H.Z. and X.H. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal