Abstract
All-trans retinoic acid (ATRA), a natural ligand for the retinoic acid receptors (RARs), induces clinical remission in most acute promyelocytic leukemia (APL) patients through the induction of differentiation and/or eradication of leukemia-initiating cells. Here, we identify a novel natural ent-kaurene diterpenoid derived from Isodon pharicus leaves, called pharicin B, that can rapidly stabilize RAR-α protein in various acute myeloid leukemic (AML) cell lines and primary leukemic cells from AML patients, even in the presence of ATRA, which is known to induce the loss of RAR-α protein. Pharicin B also enhances ATRA-dependent the transcriptional activity of RAR-α protein in the promyelocytic leukemia–RARα–positive APL cell line NB4 cells. We also showed that pharicin B presents a synergistic or additive differentiation-enhancing effect when used in combination with ATRA in several AML cell lines and, especially, some primary leukemic cells from APL patients. In addition, pharicin B can overcome retinoid resistance in 2 of 3 NB4-derived ATRA-resistant subclones. These findings provide a good example for chemical biology–based investigations of pathophysiological and therapeutic significances of RAR-α and PML-RAR-α proteins. The effectiveness of the ATRA/pharicin B combination warrants further investigation on their use as a therapeutic strategy for AML patients.
Introduction
Retinoic acid receptors (RARs) are ligand-dependent transcription factors that function in heterodimeric states involving retinoid X receptors (RXRs). Both RARs and RXRs exist in 3 isoforms, α, β and γ, and they are the master regulators of a plethora of physiological processes, such as embryo development, organ homeostasis, and cellular events such as proliferation, differentiation, and death.1,2 Almost all cases of acute promyelocytic leukemia (APL), a unique subtype of acute myeloid leukemia (AML), express oncogenic fusion protein involving the RAR-α gene due to chromosome translocations.3 A typical example is the promyelocytic leukemia (PML)–RAR-α fusion protein that is formed as a result of the t(15;17) translocation found in more than 98% of APL patients.4,5 All-trans retinoic acid (ATRA), a natural ligand for the RARs, induces complete remission in almost all PML-RAR-α–positive APL patients through the induction of terminal differentiation and/or eradication of leukemia-initiating cells (LICs) by PML-RAR-α destruction.6-8 However, the therapeutic use of ATRA is not free of concerns. Notably, the remission in patients treated with ATRA alone is of short duration because of the rapid development of retinoid resistance.9 Moreover, the clinical effectiveness of ATRA is limited to t(15;17)-positive APL patients. It is believed that one possible means to overcome these problems might be the combination of ATRA with other agents, such as arsenic trioxide (As2O3).7,10,11
On the other hand, substantial investigations suggest that RARs and RXRs are also useful drug targets for the treatment of cancer and metabolic diseases, such as diabetes and obesity.12 The success of RAR modulation in the treatment of APL has generated considerable interests in the development of RAR/RXR modulators for these diseases, as reviewed by Altucci et al.13 By screening a series of natural compounds, here we show that pharicin B, a novel natural ent-kaurene diterpenoid from Isodon pharicus leaves, can stabilize RAR-α protein in the presence and absence of ATRA and enhance the differentiation-inducing effect of ATRA in several AML cell lines and some primary leukemic cells, especially those from APL patients. Moreover, pharicin B can overcome retinoid resistance in some NB4-derived, ATRA-resistant subclones. However, pharicin B can also stabilize PML-RAR-α protein in APL cells, the catabolism of which is regarded as crucial for LIC eradication and long-term remission of mouse APL by ATRA and/or As2O3.7,14 Collectively, these findings suggest that pharicin B is an extraordinary tool to probe potential pathophysiological and therapeutic roles of RAR-α and PML-RAR-α. In addition, an approach combining ATRA with pharicin B warrants further investigation as a therapeutic strategy for myeloid leukemia.
Methods
Patients and cell lines
Bone marrow samples were collected from 12 cases of newly diagnosed AML patients at Rui-Jin and Ren-Ji Hospitals affiliated to Shanghai Jiao Tong University School of Medicine (SJTU-SM). Informed consent was obtained from all patients in accordance with the Declaration of Helsinki, and all manipulations were approved by the Medical Science Ethic Committee of SJTU-SM. Patients were diagnosed according to French-American-British classification. Detailed information of patients is listed in Table 1. Mononuclear cells were aspirated by Ficoll-Paque liquid. All primary cells, AML cell lines U937, THP-1, NB4, and NB4-derived ATRA-resistant cell lines NB4-MR2, NB4-LR1, and NB4-LR2, as well as NB4FLAG-RARα and U937FLAG-RARα cell lines with stable expression of FLAG-RAR-α (see supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) were cultured in RPMI 1640 medium (Sigma-Aldrich), supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gibco BRL), penicillin (100 IU/mL), and streptomycin (100 μg/mL) in a humidified incubator at 37°C and 5% CO2/95% air. For experiments, cells were incubated with pharicin B, ATRA (Sigma-Aldrich), AM580 (BIOMOL), Ro 41-5253 (BIOMOL), and vitamin D3 (Calbiochem), with 0.1% dimethyl sulfoxide (DMSO) treatment as control. Isolation and identification of pharicin B are shown in supplemental Methods.
Patient data and response to pharicin B and/or ATRA
| No. . | Sex . | Age, y . | FAB subtype . | Chromosome . | Blasts % . | WBC (× 109/L) . | CD11b-positive cells % . | P . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DMSO . | pharicin B . | ATRA . | pharicin B/ATRA . | * . | † . | |||||||
| 1 | F | 25 | M4 | 46, XX | 56.5 | 23.5 | 11.06 ± 0.50 | 13.29 ± 0.53 | 11.28 ± 0.58 | 15.48 ± 0.21 | > .05 | > .05 |
| 2 | M | 77 | M4 | ND | 75.5 | 52.8 | 18.72 ± 0.37 | 20.00 ± 1.69 | 26.76 ± 2.90 | 29.40 ± 3.15 | > .05 | > .05 |
| 3 | M | 33 | M5 | 46, XY | 86 | 27.7 | 1.80 ± 0.85 | 2.54 ± 0.34 | 2.32 ± 0.54 | 2.64 ± 0.17 | > .05 | > .05 |
| 4 | M | 19 | M5 | 46, XY | 80.5 | 21.8 | 10.80 ± 0.61 | 17.21 ± 1.01 | 14.02 ± 0.34 | 19.94 ± 1.77 | < .02 | > .05 |
| 5 | M | 39 | M3 | 46, XY, PML-RARα+ | 80.5 | 101 | 5.07 ± 0.37 | 5.91 ± 0.38 | 36.98 ± 0.44 | 85.12 ± 1.51 | > .05 | < .001 |
| 6 | M | 43 | M3 | 46, XY, PML-RARα+ | 94.5 | 98 | 0.46 ± 0.49 | 0.56 ± 0.27 | 38.30 ± 0.26 | 89.00 ± 0.16 | > .05 | < .001 |
| 7 | F | 36 | M4 | ND | 70.5 | 46.8 | 18.60 ± 1.14 | 24.19 ± 1.21 | 34.26 ± 3.87 | 49.17 ± 3.79 | < .02 | < .001 |
| 8 | M | 27 | M4 | ND | 83.5 | 17.9 | 23.77 ± 0.72 | 29.08 ± 0.72 | 34.25 ± 1.33 | 48.06 ± 1.00 | < .01 | < .001 |
| 9 | M | 53 | M4 | 46, XY | 79 | 69.8 | 8.53 ± 0.22 | 13.25 ± 0.65 | 33.70 ± 2.78 | 56.60 ± 1.31 | < .001 | < .001 |
| 10 | M | 61 | M5 | ND | 83.5 | 2.3 | 12.65 ± 2.62 | 26.92 ± 0.89 | 32.82 ± 2.71 | 61.92 ± 1.93 | < .001 | < .001 |
| 11 | M | 20 | M5 | ND | 77.6 | 161.2 | 15.02 ± 1.08 | 40.07 ± 0.87 | 37.38 ± 0.64 | 67.02 ± 1.46 | < .001 | < .001 |
| 12 | F | 53 | M5 | 46, XX | 92 | 58.1 | 22.47 ± 0.90 | 28.10 ± 0.98 | 40.81 ± 1.87 | 54.86 ± 2.63 | < .001 | < .002 |
| No. . | Sex . | Age, y . | FAB subtype . | Chromosome . | Blasts % . | WBC (× 109/L) . | CD11b-positive cells % . | P . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DMSO . | pharicin B . | ATRA . | pharicin B/ATRA . | * . | † . | |||||||
| 1 | F | 25 | M4 | 46, XX | 56.5 | 23.5 | 11.06 ± 0.50 | 13.29 ± 0.53 | 11.28 ± 0.58 | 15.48 ± 0.21 | > .05 | > .05 |
| 2 | M | 77 | M4 | ND | 75.5 | 52.8 | 18.72 ± 0.37 | 20.00 ± 1.69 | 26.76 ± 2.90 | 29.40 ± 3.15 | > .05 | > .05 |
| 3 | M | 33 | M5 | 46, XY | 86 | 27.7 | 1.80 ± 0.85 | 2.54 ± 0.34 | 2.32 ± 0.54 | 2.64 ± 0.17 | > .05 | > .05 |
| 4 | M | 19 | M5 | 46, XY | 80.5 | 21.8 | 10.80 ± 0.61 | 17.21 ± 1.01 | 14.02 ± 0.34 | 19.94 ± 1.77 | < .02 | > .05 |
| 5 | M | 39 | M3 | 46, XY, PML-RARα+ | 80.5 | 101 | 5.07 ± 0.37 | 5.91 ± 0.38 | 36.98 ± 0.44 | 85.12 ± 1.51 | > .05 | < .001 |
| 6 | M | 43 | M3 | 46, XY, PML-RARα+ | 94.5 | 98 | 0.46 ± 0.49 | 0.56 ± 0.27 | 38.30 ± 0.26 | 89.00 ± 0.16 | > .05 | < .001 |
| 7 | F | 36 | M4 | ND | 70.5 | 46.8 | 18.60 ± 1.14 | 24.19 ± 1.21 | 34.26 ± 3.87 | 49.17 ± 3.79 | < .02 | < .001 |
| 8 | M | 27 | M4 | ND | 83.5 | 17.9 | 23.77 ± 0.72 | 29.08 ± 0.72 | 34.25 ± 1.33 | 48.06 ± 1.00 | < .01 | < .001 |
| 9 | M | 53 | M4 | 46, XY | 79 | 69.8 | 8.53 ± 0.22 | 13.25 ± 0.65 | 33.70 ± 2.78 | 56.60 ± 1.31 | < .001 | < .001 |
| 10 | M | 61 | M5 | ND | 83.5 | 2.3 | 12.65 ± 2.62 | 26.92 ± 0.89 | 32.82 ± 2.71 | 61.92 ± 1.93 | < .001 | < .001 |
| 11 | M | 20 | M5 | ND | 77.6 | 161.2 | 15.02 ± 1.08 | 40.07 ± 0.87 | 37.38 ± 0.64 | 67.02 ± 1.46 | < .001 | < .001 |
| 12 | F | 53 | M5 | 46, XX | 92 | 58.1 | 22.47 ± 0.90 | 28.10 ± 0.98 | 40.81 ± 1.87 | 54.86 ± 2.63 | < .001 | < .002 |
ND indicates not done.
P value between pharicin B and DMSO.
P value between pharicin B/ATRA and pharicin B or ATRA treatment alone
Cell cycle and differentiation assay
The distribution of nuclear DNA contents was analyzed by flow cytometry (FACSCalibur; BD Biosciences). Cell morphological features were examined by microscopy after Wright's staining (BASO Diagnostic) and cell-surface differentiation antigens CD11b and CD14 were measured using fluorescein isothiocyanate– or phycoerythrin-labeled antibodies with isotype controls (Beckman-Coulter) via flow cytometry. The nitroblue tetrazolium chloride (NBT) reduction was performed as previously described,15 and 200 cells on each slide were counted under a light microscope (Olympus BX-51; Olympus Optical) and the percentage of NBT-positive cells was calculated.
Pulse-chase assay
NB4FLAG-RARα cells were preincubated in cysteine/methionine-free RPMI 1640 medium for 1 hour and then pulse-labeled with 50 μCi/mL [35S]methionine/cysteine (Perkin-Elmer) for 1 hour. After washing with phosphate-buffered saline (PBS) twice, cells were chased with an excess amount of cold cysteine/methionine in serum supplemented normal medium with or without 2μM pharicin B. Equal amounts of cells were harvested at different time points, and cells were lysed with radioimmune precipitation buffer (50mM Tris, 150mM NaCl, 1mM EGTA [ethylene glycol bis(B-aminoethyl ether)-N,N,N′,N′-tetraacetic acid], 1mM deoxycholic acid, 1% NP40, pH 7.4), plus a protease-inhibitor cocktail (Roche Diagnostic). Whole-cell extracts were incubated with anti-FLAG M2 resin (Sigma-Aldrich) overnight at 4°C. After resins were washed 3× with radioimmune precipitation buffer, samples were eluted with sodium dodecyl sulfate (SDS) and then subjected to 10% SDS-polyacrylamide gel electrophoresis (PAGE). Gels were dried and then visualized by STORM PhosphorImager (Molecular Dynamics). The signal intensity of the bands was quantified using Quantity One Version 4.4.0 (Bio-Rad).
ChIP
After crosslinking with 1% formaldehyde at room temperature for 10 minutes, cells were pelleted and resuspended in 400 μL lysis buffer (1% SDS, 10mM EDTA [ethylenediaminetetraacetic acid], 50mM Tris-HCl, pH 8.0) plus protease-inhibitor cocktail. Cellular DNA was then sonicated and sheared to small fragments of 500-1000 bp with a Sonicator ultrasonic processor (Misonix Inc). Subsequently, the supernatant of the sonicated cells was collected, diluted, and precleared by protein A agarose (Santa Cruz Biotechnology). Furthermore, anti-FLAG M2 affinity resin (Sigma-Aldrich) was added to the supernatant for immunoprecipitation. After incubation overnight, resins were washed with low-salt, high-salt, and LiCl buffers, and the immunoprecipitated DNA was retrieved by 5M NaCl at 65°C for 4 hours and then purified with a polymerase chain reaction (PCR) purification kit (QIAGEN). PCR for the retinoic acid response element (RARE) in the promoter of RAR-β was performed with specific primers 5′-TCCTGGGAGTTGGTGATGTCAG-3′ (forward) and 5′-AAACCCTGCTCGGA TCGCTC -3′ (reverse).
Real-time quantitative PCR
For quantitative analysis of gene expression, total RNA was isolated by a Trizol kit (Invitrogen). RNA was treated with DNase (Promega). Complementary DNA was synthesized according to the manufacturer's instruction (Promega). Real-time quantitative PCRs for RAR-β, CCAAT/enhancer binding protein-beta (C/EBP-β), retinoic acid–induced genes E, I, and G (RIG-E, RIG-I, and RIG-G), interferon regulatory factor 1 (IRF-1), transglutaminase 2 (TGM2), ubiquitin-activating enzyme-E1–like protein (UBE1L), and myeloperoxidase (MPO) were performed with SYBR Green PCR Master Mixture Reagents (Applied Biosystems) on the ABI PRISM 7900 system (Applied Biosystems). The specific primers used for detecting genes are listed in supplemental Table 1. PCR was initiated with a step of uracil-N-glycosylase incubation at 50°C for 2 minutes to remove DNA contamination, followed by 95°C for 10 minutes to hot-start the DNA polymerase and denature the template, and then 40 cycles consisted of denaturing at 95°C for 30 seconds, annealing at 58°C for 30 seconds, and extension at 72°C for 30 seconds. Data were analyzed as previously reported.16
Western blot
Cell lysates were equally loaded onto 10% SDS-PAGE, electrophoresed, and transferred to enhanced chemiluminescence–nitrocellulose membranes (Amersham). The membranes were stained with 0.2% Ponceau S red to ensure equal protein loading. After blocking with 5% nonfat milk in Tris-buffered saline, the membranes were incubated with the polyclonal antibodies against RAR-α (SC-551; Santa Cruz Biotechnology), RXR-α, inhibitor of kappa B alpha (IκBα), vitamin D receptor (VDR), FLAG (Santa Cruz Biotechnology), c-Myc (Cell Signaling Technology), and neuregulin receptor degradation protein-1 (Nrdp1; Bethyl Laboratories) overnight at 4°C, followed by horseradish peroxidase (HRP)–linked secondary antibody (Cell Signaling) for 1 hour at room temperature. Detection was performed by a SuperSignal West Pico Chemiluminescent Substrate kit (Pierce), according to the manufacturer's instruction. β-actin (Merck) was used as an internal control. Signal intensity of proteins was normalized against β-actin using Quantity One (Bio-Rad).
Statistical analysis
Student t test was used to evaluate the difference between 2 different treatments. A P value of less than .05 was considered statistically significant.
Results
Pharicin B stabilizes steady-state RAR-α protein level in AML cells
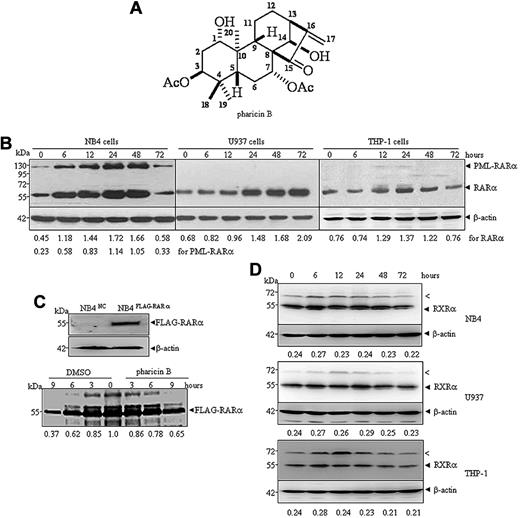
Pharicin B is a novel natural ent-kaurene diterpenoid with the molecular formula C24H34O7 (Figure 1A) that was isolated from the leaves of I pharicus (a kind of Chinese herbal plant found mainly in Tibet), as described in supplemental Methods. Three AML cell lines (NB4, U937, and THP-1 cells) were treated with different concentrations of pharicin B to determine its potential cytotoxicity. The results showed that the natural compound exerted dose-dependent growth inhibition on all 3 cell lines to a similar degree (supplemental Figure 1A). Pharicin B at a concentration of 4μM or higher significantly decreased cell viability, while more than 90% cells treated with pharicin B at 1 and 2μM remained alive (supplemental Figure 1B). In the subsequent experiments, we mainly screened the potential effects of the nontoxic concentrations of pharicin B on these AML cells. Interestingly, 2μM of pharicin B rapidly and significantly increased steady-state RAR-α protein level (Figure 1B), but not its mRNA level (supplemental Figure 1C), in all 3 AML cell lines tested. A relatively weaker effect on RAR-α was seen in THP-1 cells, compared with that in NB4 and U937 cells. In addition, pharicin B could accumulate ectopically expressed FLAG-RAR-α protein (data not shown). To detect whether pharicin B influences the translation and/or degradation of RAR-α protein, a pulse-chase experiment was performed in NB4 cells with stable ectopic expression of FLAG-tagged RAR-α (Figure 1C top panel) due to the unavailability of an effective anti–RAR-α antibody for immunoprecipitation. As depicted in the bottom panel of Figure 1C, 65.12% of isotope-labeled RAR-α protein remained in cells with pharicin B treatment for 9 hours, while only 36.65% of RAR-α could be seen in untreated cells, suggesting that pharicin B may stabilize RAR-α proteins. Interestingly, APL cell-specific PML-RAR-α protein also increased in the same dynamic manner as wild-type RAR-α protein in pharicin B–treated APL cell line NB4 cells (Figure 1B). It was worth noting that when long incubation time (72 hours) was allowed, increased RAR-α and/or PML-RAR-α protein was decreased in NB4 or THP-1 cells. We extrapolated that this was possibly due to the cell context–dependent intracellular stability of pharicin B, which remained to be shown. To test the potential specificity for RAR-α stabilization, we also tested other several proteins, including RXR-α (Figure 1D), VDR, IκBα, c-Myc, and Nrdp1 (supplemental Figure 2) with pharicin B treatment. Nrdp1 was reported to act as E3 ligase to regulate the differentiation of hematopoietic progenitors by governing steady-state levels of cytokine and RARs.17 Our results demonstrated that except for c-Myc that was decreased under treatment of pharicin B, the compound had no significant effects on RXR-α (Figure 1D), VDR, IκBα, and Nrdp1 protein in 3 AML cell lines tested (supplemental Figure 2). Notably, pharicin B appeared to transiently stabilize an anti–RXR-α antibody-responsive protein with approximately 70 kDa (Figure 1D the arrowed “<” band), which was not sumoylated RXR-α protein because SUMO1 or SUMO2/3 could not be detected in anti–RXR-α antibody–immunoprecipitated complex at 70 kDa in pharicin B–treated or –untreated NB4 cells (data not shown).
Pharicin B stabilizes RAR-α protein in AML cells. (A) Chemical structure of pharicin B. (B) NB4, U937, or THP-1 cells were incubated with 2μM of pharicin B for hours, as indicated. The RAR-α/PML-RAR-α protein was detected by anti–RAR-α antibody with β-actin as the loading control. The number on the bottom indicates signal intensity of RAR-α or PML-RAR-α protein against β-actin. (C) NB4 cells were stably transfected with FLAG-RAR-α or the negative control (NC) vector (top panel). Pulse-chase analysis of catabolism of FLAG-RAR-α in the presence or absence of pharicin B was performed in NB4FLAG-RARα cells (bottom panel). The number on the bottom indicates the signal intensity of the FLAG-RAR-α relative to untreated cells, which was designated as 1. (D) NB4, U937, or THP-1 cells were incubated with 2μM of pharicin B for the number of hours indicated, and RXR-α protein was detected by anti–RXR-α antibody with β-actin as the loading control. The number on the bottom indicates signal intensity of RXR-α protein against β-actin. The arrow (<) points to an unknown band responsive to anti–RXR-α antibody. All experiments were repeated 3 times.
Pharicin B stabilizes RAR-α protein in AML cells. (A) Chemical structure of pharicin B. (B) NB4, U937, or THP-1 cells were incubated with 2μM of pharicin B for hours, as indicated. The RAR-α/PML-RAR-α protein was detected by anti–RAR-α antibody with β-actin as the loading control. The number on the bottom indicates signal intensity of RAR-α or PML-RAR-α protein against β-actin. (C) NB4 cells were stably transfected with FLAG-RAR-α or the negative control (NC) vector (top panel). Pulse-chase analysis of catabolism of FLAG-RAR-α in the presence or absence of pharicin B was performed in NB4FLAG-RARα cells (bottom panel). The number on the bottom indicates the signal intensity of the FLAG-RAR-α relative to untreated cells, which was designated as 1. (D) NB4, U937, or THP-1 cells were incubated with 2μM of pharicin B for the number of hours indicated, and RXR-α protein was detected by anti–RXR-α antibody with β-actin as the loading control. The number on the bottom indicates signal intensity of RXR-α protein against β-actin. The arrow (<) points to an unknown band responsive to anti–RXR-α antibody. All experiments were repeated 3 times.
Pharicin B accumulates RAR-α protein in the presence of ATRA in AML cell lines and primary cells
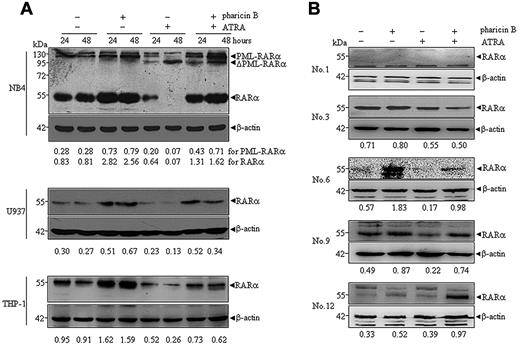
As previously documented,18,19 ATRA at pharmacological concentrations induced a progressive degradation of wild-type RAR-α in NB4, U937, and THP-1 cells, and also cleaved PML-RAR-α protein into a 90-kDa product (ΔPML-RAR-α) in NB4 cells (Figure 2A). When these cells were cotreated with ATRA and pharicin B for 24 and 48 hours, more intriguingly, wild-type RAR-α protein levels still remained high, especially in NB4 and U937 cells (Figure 2A). In addition, pharicin B did not appear to block ATRA-induced cleavage of PML-RAR-α protein, but it could increase the amount of full-length PML-RAR-α in NB4 cells (Figure 2A), suggesting that pharicin B also stabilized PML-RAR-α protein with no influence on ATRA-induced PML-RAR-α cleavage. Furthermore, we checked the expression of RAR-α protein in some AML patients who had enough cells available for Western blotting. As shown in Figure 2B, pharicin B stabilized RAR-α protein in the presence and absence of ATRA in patients 6, 9, and 12. Patient 1 was deficient in RAR-α expression, and no changes of RAR-α protein were observed in patient 3.
Pharicin B accumulates RAR-α protein in the presence of ATRA in AML cell lines and some primary cells. Cells were treated with pharicin B (2μM) and/or ATRA (10−8M for NB4 and 10−7M for U937 and THP-1) for 24 and 48 hours (A), and primary AML cells isolated from newly diagnosed patients (number represents patient numbers listed in Table 1) were treated with 2μM pharicin B and/or 10−7M ATRA for 48 hours (B). The RAR-α/PML-RAR-α protein was detected by anti–RAR-α antibody with β-actin as the loading control. The number on the bottom indicates signal intensity of RAR-α or PML-RAR-α protein against β-actin.
Pharicin B accumulates RAR-α protein in the presence of ATRA in AML cell lines and some primary cells. Cells were treated with pharicin B (2μM) and/or ATRA (10−8M for NB4 and 10−7M for U937 and THP-1) for 24 and 48 hours (A), and primary AML cells isolated from newly diagnosed patients (number represents patient numbers listed in Table 1) were treated with 2μM pharicin B and/or 10−7M ATRA for 48 hours (B). The RAR-α/PML-RAR-α protein was detected by anti–RAR-α antibody with β-actin as the loading control. The number on the bottom indicates signal intensity of RAR-α or PML-RAR-α protein against β-actin.
Pharicin B enhances ATRA-dependent transcriptional activity of RAR-α protein in PML-RAR-α–positive NB4 cells
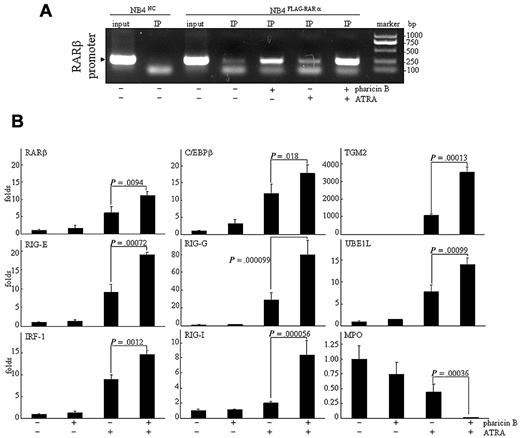
The association of the RAR/RXR heterodimer with specific DNA sequences, called the RAREs, in the promoter regions of target genes is essential for the ligand (ATRA)-dependent modulation of gene expression.20 We used the chromatin immunoprecipitation (ChIP) assay to scrutinize whether pharicin B–stabilized RAR-α protein would bind to these regions with RARE ex vivo. For this purpose, NB4FLAG-RARα cells were treated with pharicin B and/or ATRA, and then FLAG-RAR-α protein was precipitated with anti-FLAG antibody. As can be seen in Figure 3A, FLAG-RAR-α protein stabilized by pharicin B effectively bound to RARE located at the promoter of RAR-β (a known target gene of RAR-α21 ), and the binding ability of RAR-α protein was significantly enhanced by the pharicin B/ATRA combination. Previous investigations proposed that the ligand-dependent recruitment of chromatin-remodeling activity serves as a general mechanism underlying the switch of nuclear receptors from being transcriptionally repressive to being transcriptionally active.22 Therefore, the increased RAR-α binding does not mean that there was increased transcriptional activity of RAR-α protein. Consistent with previous reports,21,23-27 the ATRA, but not pharicin B, treatment increased RAR-β, C/EBPβ, RIG-E, RIG-I, RIG-G, IRF-1, TGM2, and UBE1L gene expression and decreased that of MPO in NB4 (Figure 3B), U937, and THP-1 cells (supplemental Figure 3). More intriguingly, the modulation of these expressions by ATRA was significantly enhanced by the cotreatment with pharicin B in NB4 cells (Figure 3B), but not in U937 and THP-1 cells, in which only increased RIG-G could be seen with pharicin B/ATRA cotreatment (supplemental Figure 3).
Pharicin B enhances ATRA-regulated gene expression in NB4 cells. (A) NB4FLAG-RARα cells were treated as indicated, and ChIP experiment was performed as described in the Methods. NB4NC cells were used as the negative control. (B) NB4 cells were treated by 2μM pharicin B and/or 10−8M ATRA for 48 hours, and the expression of genes, as indicated, were detected by real-time PCR with specific primers. P values between pharicin B plus ATRA and ATRA are shown. Each experiment was repeated 3 times.
Pharicin B enhances ATRA-regulated gene expression in NB4 cells. (A) NB4FLAG-RARα cells were treated as indicated, and ChIP experiment was performed as described in the Methods. NB4NC cells were used as the negative control. (B) NB4 cells were treated by 2μM pharicin B and/or 10−8M ATRA for 48 hours, and the expression of genes, as indicated, were detected by real-time PCR with specific primers. P values between pharicin B plus ATRA and ATRA are shown. Each experiment was repeated 3 times.
Pharicin B enhances differentiation induction of ATRA in AML cell lines and primary blasts from some AML patients
Next, we detected the cellular effect of nontoxic concentrations of pharicin B on the AML cell lines, NB4, U937, and THP-1, with 10−6 M of ATRA or with 10−7 M of vitamin D3 as a positive control. Pharicin B inhibited cell proliferation in a time-dependent manner, with an increased percentage of cells at the G1 stage (supplemental Figure 4A-B). Compared with the untreated and ATRA-treated cells, pharicin B–treated cells presented a slightly decreased nuclear/cytoplasm ratio and cell size and a low degree of CD11b+/CD14− expression without NBT reduction (supplemental Figure 4). We presumed that the slight differentiation-inducing effects of pharicin B alone was due to the existence of the physiological concentration of retinoic acid in the FBS. Thus, pharicin B and low concentration (10−8M) of ATRA were applied individually or in combination to NB4 cells. As depicted in Figure 4, the pharicin B and ATRA combination synergistically induced growth arrest and terminal granulocytic differentiation, as evidenced by mature granulocytic morphologic features (such as smaller cell size, reduced nucleus-cytoplasm ratio, condensed chromatin, distorted, stab form, and lobed nuclei) and increased NBT reduction and percentage of CD11b+ cells, with the latter being free of CD14 (supplemental Figure 5A), a monocytic differentiation marker.28 Such a synergistic effect exhibited in the use of ATRA and pharicin B in differentiation and growth inhibition could also be seen in the leukemic U937 and THP-1 cells (supplemental Figure 5). Furthermore, we also determined the potential differentiation-inducing effects of pharicin B at 2μM and/or ATRA at 10−7 M on primary blast cells from 12 cases of AML patients, according to the percentage of CD11b-positive cells (Table 1) and morphologic criteria (supplemental Figure 6). Although the number of cases was small, the patients fell into 3 classes, according to their responsiveness to ATRA and pharicin B, excluding patients 1-4, who presented no response to ATRA and ATRA/pharicin B combination: (1) no basal effect of pharicin B with sensitiveness to ATRA, but major synergy with ATRA (patients 5 and 6, who suffered from APL); (2) clear effect of pharicin B and additive effect with ATRA (patients 10 and 11), and (3) low sensitivity to pharicin B, but synergy with ATRA (patients 7, 8, 9, and 12).
Pharicin B enhances the differentiation-inducing effect of ATRA in NB4 cells. (A-B) NB4 cells were incubated with 1 or 2μM pharicin B and/or 10−8M ATRA for the indicated days, and viable cells were determined by trypan-blue exclusion assay (A), and CD11b-positive cells were counted by flow cytometry (B). *P < .001 between pharicin B/ATRA- and ATRA-treated cells. (C-D) The morphology (C) and NBT staining (D) during the 3-day treatment are indicated. In panel D, the values represent means ± SD) of NBT-positive percentages of triplicate samples. All experiments were repeated 3 times.
Pharicin B enhances the differentiation-inducing effect of ATRA in NB4 cells. (A-B) NB4 cells were incubated with 1 or 2μM pharicin B and/or 10−8M ATRA for the indicated days, and viable cells were determined by trypan-blue exclusion assay (A), and CD11b-positive cells were counted by flow cytometry (B). *P < .001 between pharicin B/ATRA- and ATRA-treated cells. (C-D) The morphology (C) and NBT staining (D) during the 3-day treatment are indicated. In panel D, the values represent means ± SD) of NBT-positive percentages of triplicate samples. All experiments were repeated 3 times.
Pharicin B presents synergistic effect with RAR-α agonist, while RAR-α antagonist inhibits the differentiation-enhancing effect of pharicin B on ATRA
To further confirm the role of the stabilized RAR-α protein in the synergistic differentiation-inducing effect of the pharicin B plus ATRA combination, AM580, a selective RAR-α agonist,29 was applied to NB4 and U937 cells. The results showed that AM580 induced differentiation of NB4 and U937 cells in a concentration-dependent manner, as assessed by CD11b expression (supplemental Figure 7A) and differentiation-related morphological criteria (supplemental Figure 7B). Notably, AM580-induced differentiation was more significant in NB4 than in U937 cells. Like that seen under ATRA, AM580-induced differentiation of NB4 and U937 cells could also be significantly enhanced by pharicin B (supplemental Figure 7A-B). In addition, pharicin B could accumulate PML-RAR-α and/or RAR-α protein in the presence of AM580 in NB4 and U937 cells, although AM580-only treatment induced a loss of these 2 proteins (Figure 5A). On the other hand, Ro 41-5253, a highly specific RAR-α antagonist,30 could effectively antagonize the synergistic differentiation-inducing effect of the pharicin B and ATRA combination in U937 and, especially, in NB4 cell lines (Figure 5B and supplemental Figure 7C).
Pharicin B presents synergistic effect with RAR-α agonist, while RAR-α antagonist inhibits effect of pharicin B. (A) NB4 and U937 cells were treated with 2μM pharicin B, ATRA (10−8M for NB4 and 10−7M for U937), or AM580 (10−6M), as indicated, for 48 hours, and RAR-α/PML-RAR-α protein was detected by anti–RAR-α antibody. β-actin was used as the internal control. Each experiment was repeated at least 3 times. The number on the bottom indicates signal intensity of RAR-α or PML-RAR-α protein against β-actin. (B) NB4 and U937 cells were treated with pharicin B (2μM), ATRA (10−8M for NB4 and 10−7M for U937), 10−6M Ro 41–5253 alone, or their combination for 3 days. CD11b expression was measured by flow cytometry. All values represent means, with bar as SD of 3 independent experiments, each of which with triplicate samples. *P < .001.
Pharicin B presents synergistic effect with RAR-α agonist, while RAR-α antagonist inhibits effect of pharicin B. (A) NB4 and U937 cells were treated with 2μM pharicin B, ATRA (10−8M for NB4 and 10−7M for U937), or AM580 (10−6M), as indicated, for 48 hours, and RAR-α/PML-RAR-α protein was detected by anti–RAR-α antibody. β-actin was used as the internal control. Each experiment was repeated at least 3 times. The number on the bottom indicates signal intensity of RAR-α or PML-RAR-α protein against β-actin. (B) NB4 and U937 cells were treated with pharicin B (2μM), ATRA (10−8M for NB4 and 10−7M for U937), 10−6M Ro 41–5253 alone, or their combination for 3 days. CD11b expression was measured by flow cytometry. All values represent means, with bar as SD of 3 independent experiments, each of which with triplicate samples. *P < .001.
Pharicin B restores ATRA sensitivity in some APL cell–derived ATRA-resistant cells
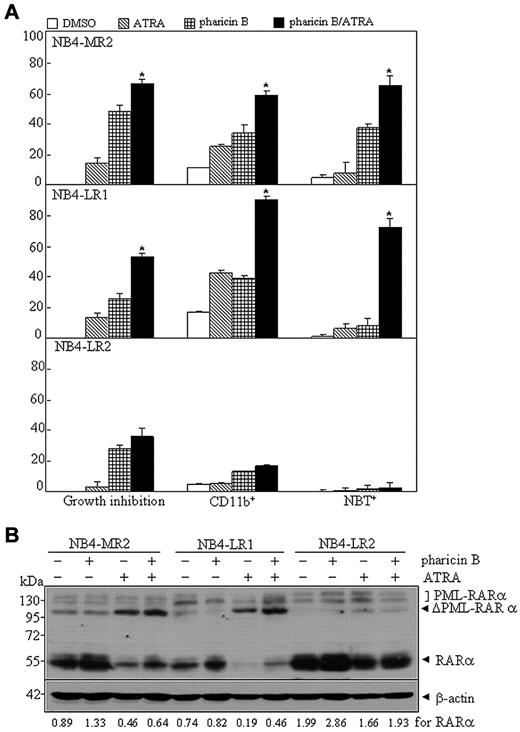
Many ATRA-resistant subclones of the human APL cell line, NB4, were established, despite the fact that the exact mechanisms to their retinoid resistance were largely unknown.31-33 For example, NB4-LR1 is an ATRA-responsive cell line in which ATRA alone fails to induce maturation, but renders these cells competent to a cyclic AMP (cAMP)–signaling-triggered differentiation.31 Here, we tested whether the pharicin B/ATRA (10−7M) combination could cause differentiation in 3 NB4-derived subclones, including NB4-MR2, NB4-LR1, and NB4-LR2.31,32 In accordance with previous reports,11,31,34 these 3 subclones were refractory to ATRA-induced maturation, as assessed by growth inhibition, CD11b expression, NBT-reducing activity (Figure 6A), and morphological criteria (supplemental Figure 8). The single treatment of pharicin B caused a growth inhibition in all 3 cell lines and induced differentiation, to a low degree, in NB4-MR2 cells, but not in NB4-LR1 and NB4-LR2 (Figure 6A and supplemental Figure 8). However, administration of pharicin B along with ATRA significantly increased CD11b-positive cells, NBT response, and differentiation-related morphological features in NB4-MR2 and NB4-LR1, but not in NB4-LR2, cells (Figure 6A). More intriguingly, pharicin B alone could accumulate RAR-α protein in NB4-MR2 and NB4-LR2, and ATRA induced a degradation of RAR-α protein in NB4-MR2 and NB4-LR1. However, unlike that seen in ATRA-sensitive NB4 cells, the pharicin B/ATRA combination induced a stabilization of RAR-α protein, to a lesser degree (Figure 6B). In addition, NB4-LR2 subclone was reported to express the wild-type PML-RAR-α transcript, in addition to the transcript harboring a point mutation in the retinoid-binding domain of PML-RAR-α that generates an in-frame termination codon, leading to the deletion of 52 amino acids at its C-terminal end.35 However, it was reported to be undetectable with anti–RAR-α or anti-PML antibody, presumably due to its instability.36 Here, we could detect PML-RAR-α protein in NB4-LR2 cells with anti–RAR-α antibody (SC-551; Santa Cruz Biotechnology; Figure 6B). This difference from the previous report36 might have been that different origin of antibody was used in these works. ATRA could also cleave PML-RAR-α protein in all 3 resistant subclones, especially in NB4-MR2 and NB4-LR1 cells, which was not affected by the presence of pharicin B (Figure 6B).
Pharicin B overcomes ATRA resistance in some NB4-derived and ATRA-resistant cell lines. (A) Percentages of growth inhibition, CD11b expression, and NBT reduction for ATRA-resistant cells NB4-MR2, NB4-LR1, and NB4-LR2 treated with 2μM pharicin B, 0.1μM ATRA, or both for 3 days. (B) Cells were treated as in (A), but for 48 hours, and RAR-α/PML-RAR-α protein was detected by antibody of RAR-α. β-actin was used as the internal control. Each experiment was repeated at least 3 times. The number on the bottom indicates signal intensity of RAR-α protein against β-actin.
Pharicin B overcomes ATRA resistance in some NB4-derived and ATRA-resistant cell lines. (A) Percentages of growth inhibition, CD11b expression, and NBT reduction for ATRA-resistant cells NB4-MR2, NB4-LR1, and NB4-LR2 treated with 2μM pharicin B, 0.1μM ATRA, or both for 3 days. (B) Cells were treated as in (A), but for 48 hours, and RAR-α/PML-RAR-α protein was detected by antibody of RAR-α. β-actin was used as the internal control. Each experiment was repeated at least 3 times. The number on the bottom indicates signal intensity of RAR-α protein against β-actin.
Discussion
The antitumor activities of some small molecules identified from Isodon plants, which are mainly distributed in southwest China, have been closely examined. For example, oridonin and eriocalyxin B exert antileukemia activity through inducing apoptosis.37,38 In this work, we provided the first demonstration that when nontoxic concentrations of pharicin B were applied to ATRA-sensitive leukemic cells, wild-type RAR-α protein, but not its mRNA, was accumulated. Such an accumulation could also be seen in ectopically expressed RAR-α protein. Although we could not exclude the possibility that pharicin B could also affect the translation of RAR-α protein, the pulse-chase experiment showed that pharicin B could effectively stabilize RAR-α protein. Furthermore, pharicin B also stabilized the PML-RAR-α fusion protein, but did not affect its cleavage by ATRA in NB4 cells. Previous studies showed ATRA-triggered degradation of both RAR-α and PML-RARα proteins was mediated by the ubiquitin-proteasome pathway.19,39 The fact that pharicin B accumulated RAR-α protein in the presence of ATRA in AML cell lines and some primary cells from some AML patients suggested that pharicin B stabilized RAR-α protein through a mechanism independent from ATRA-induced loss of RAR-α protein. Therefore, it remains to be further explored how pharicin B stabilizes RAR-α protein.
Although PML-RAR-α was shown to exert dominant-negative effects on the activity of wild-type RAR-α protein,40 exogenous expression of PML-RAR-α in U937 cells increased ATRA sensitivity,41 and the enhanced expression of PML-RAR-α in the ATRA-resistant NB4.007/6 cells was sufficient to restore an ATRA-sensitive phenotype.42 Recently, ATRA was proposed to induce a switch in PML-RAR-α activity from a repressor to an activator of transcription, which set off a wave of differentiation events and thus the elimination of a large fraction of malignant cells and PML-RAR-α itself.7,43 Indeed, we showed that, with the stabilization of RAR-α protein, pharicin B enhanced the ATRA-dependent transcriptional activity of RAR-α protein in PML-RAR-α–positive NB4 cells. Except for the increased RIG-G expression, however, such an enhancement could not be seen in U937 and THP-1 cells, consistent with the previous observation that ATRA could mediate a direct increase of C/EBP-β only in PML-RAR-α–expressing cells.44
On the other hand, some reagents capable of preventing ATRA-induced loss of RAR-α and/or PML-RAR-α protein have been shown to enhance ATRA-induced differentiation.45-48 Hence, we investigated whether pharicin B could enhance the differentiation-inducing activity of ATRA. Although only a slight differentiation effect was observed for the pharicin B–only treatment, pharicin B could synergistically increase ATRA- as well as RAR-α agonist AM580-induced terminal differentiation in AML cell lines, which was, in turn, inhibited by an RAR-α antagonist. Such a synergistic or additive action of pharicin B could also be seen in some primary AML cells, consistent with the stabilization of RAR-α protein. All of these data indicated that maintenance of RAR-α protein contributed to the enhancing effect of pharicin B on ATRA-induced differentiation. However, pharicin B induced a significant increase in G1 arrest (supplemental Figure 4B) and in CD11b expression (supplemental Figure 4D) in THP-1 cells, while stabilization of RAR-α was not as significant in this cell line as seen in NB4 and U937 cells (Figure 1B). Remarkably, the differentiation-enhancing effect of pharicin B with ATRA was not paralleled to its RAR-α–stabilized effect in NB4-derived, ATRA-resistant cell lines, especially in NB4-MR2. These results suggested that, in addition to stabilization of RAR-α, other factors may also contribute to the differentiation-enhancing effect of pharicin B on ATRA. In fact, we showed that pharicin B could also decrease the expression of c-Myc protein, which has been shown to contribute to differentiation induction in AML cells.49 Furthermore, agents that block the cell cycle were known to nonspecifically enhance ATRA-induced differentiation.50,51 Therefore, it also deserves to be considered that the differentiation-enhancing effect of pharicin B on ATRA may be related with its growth-arresting effect.
Recent investigations showed that PML-RAR-α alone can confer self-renewal to committed hematopoietic progenitors before the onset of disease.52 A relationship between the capacity of ATRA and, especially, As2O3, to target LICs, also known as leukemic stem cells or leukemia-repopulating cells,8 and their ability to induce prolonged relapse-free survival were also explored.14 Nasr et al.7 proposed that the effectiveness of ATRA in APL is independent of their ability to cause differentiation, while targeted destruction of the PML-RAR-α oncoprotein appears to be the key to eliminating LICs and inducing long-term remission of mouse APL. Here, we showed that pharicin B stabilized PML-RAR-α protein, although it did not affect cleavage of the fusion oncoprotein induced by ATRA. Considering these discoveries, the in vivo therapeutic effects and influences on LICs of pharicin B and its combination with ATRA remain to be explored. To this end, chemical synthesis of pharicin B should be considered so as to get enough of the quantity of compound for its in vivo investigation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We appreciate Dr Han-Yi Zhuang for her reading and editing. Z.G. is a PhD candidate at the Shanghai Institutes for Biological Sciences of Chinese Academy of Sciences, and this work was submitted in partial fulfillment of the requirement for the PhD.
This work was supported, in part, by grants from the National Basic Research Program of China (973 Program; NO2009CB918404, NO2010CB912104), the National Natural Science Foundation of China (NSFC; 30630034, 30870523, and 90813034), The NSFC-joint Foundation of Yunnan Province (U0832602), the Chinese Academy of Sciences (KSCX2-YW-R-097), and the Science and Technology Commission of Shanghai (08JC1413400, 08431900700). G-Q.C. was supported by the Shanghai Ling-Jun Talent Program.
Authorship
Contribution: Z.-M.G. and Y.-L.W. performed most experiments and drafted the article; M.-Y.Z. and C.-X.L. performed some experiments to evaluate differentiation; Y.Z. and H.-D.S. contributed to the isolation and identification of pharicin B; H.-Z.X. and Y.H. performed experiments on ChIP and fresh AML cells; H.Y. collected samples and analyzed clinical data from patients; and G.-Q.C. conceived and designed the study, critically revised the manuscript for important intellectual content, and approved the version to be published.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Guo-Qiang Chen, Shanghai Jiao Tong University School of Medecine, No. 280, Chong-Qing South Rd, Shanghai 200025, China; e-mail: chengq@shsmu.edu.cn.
References
Author notes
Z-M.G. and Y-L.W.contributed equally to this work.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal