Abstract
Genomic aberrations are of predominant importance to the biology and clinical outcome of patients with acute myelogenous leukemia (AML), and conventional karyotype-based risk classifications are routinely used in clinical decision making in AML. One of the known limitations of cytogenetic analysis is the inability to detect genomic abnormalities less than 5 Mb in size, and it is currently unclear whether overcoming this limitation with high-resolution genomic single-nucleotide polymorphism (SNP) array analysis would be clinically relevant. Furthermore, given the heterogeneity of molecular mechanisms/aberrations that underlie the conventional karyotype-based risk classifications, it is likely that further refinements in genomic risk prognostication can be achieved. In this study, we analyzed flow cytometer–sorted, AML blast-derived, and paired, buccal DNA from 114 previously untreated prospectively enrolled AML patients for acquired genomic copy number changes and loss of heterozygosity using Affymetrix SNP 6.0 arrays, and we correlated genomic lesion load and specific chromosomal abnormalities with patient survival. Using multivariate analyses, we found that having ≥ 2 genomic lesions detected through SNP 6.0 array profiling approximately doubles the risk of death when controlling for age- and karyotype-based risk. Finally, we identified an independent negative prognostic impact of p53 mutations, or p53 mutations and 17p-loss of heterozygosity combined on survival in AML.
Introduction
It is well established that cytogenetics and mutations in certain genes are of predominant importance to the biology and clinical outcome of patients with acute myelogenous leukemia (AML).1-18 Consequently, clinical management and decision making in AML relies heavily on risk categorization based on conventional karyotyping, a result of a decades-long series of correlations between karyotypes and survival outcomes in AML.
One of the contemporary questions regarding genomic risk prediction based on conventional karyotyping relates to the fact that small losses or gains (< 5 Mb) are not detectable and therefore whether the reliable detection of such lesions is clinically relevant. In addition, acquired states of acquired uniparental disomy (aUPD, often associated with mutated genes9 ), are not detectable using conventional karyotyping, but may have effects on AML outcome. Furthermore, despite a known association of p53 mutations with a fraction of complex karyotype AML and poor outcome, a mechanistic understanding of the relationship between other karyotyping results demonstrating genomic imbalances and AML outcome is incomplete.19,20 Conversely, genomic array-based analyses usually do not detect balanced chromosomal translocations (unless suggested by chromosomal material lost at the junctions) and are not very sensitive to clonal changes that occur in < 25% of cells.21 Therefore, it is possible that a more accurate, complete description of genomic copy number aberrations and specific genomic aberrations in AML could further refine risk prognostication, and that conventional karyotyping and high-resolution, array-based karyotyping may have complementary effects on risk prognostication in AML.
Efforts at mapping subchromosomal genomic copy-number changes using intermediate-resolution, array-based comparative genomic hybridization in AML have identified novel genomic losses and gains, and candidate genes have been proposed.22-26 Recent developments in genome-wide, high-resolution copy-number and loss of heterozygosity (LOH) analysis using single-nucleotide polymorphism (SNP) arrays have aided better definitions of the pathologic anatomy of cancer genomes, and application of SNP array technology to hematologic cancers has refined the knowledge of the anatomy of clinically important chromosomal lesions.27-35 In AML, a series of SNP array-based genomic studies has improved the characterization of the genomic lesions in AML, but few studies have correlated ultra-high-resolution, SNP array-based genomics in AML with survival outcome.26,29,36-39
For this study, we used ultra-high-resolution Affymetrix SNP 6.0 arrays to comprehensively interrogate the genomes of a large panel of previously untreated adult AML cases for subchromosomal deletions, gains and amplifications, microdeletions, aUPD, monosomies, and multisomies. Through subsequent correlative analysis of SNP array-defined genomic complexity in AML, as well as individual SNP array-defined genomic aberrations with survival, we were able to demonstrate that ultra-high-resolution, SNP array-based genomic lesion analysis adds independent negative prognostic information to age and conventional karyotyping. Furthermore, in this study, we identified an independent negative prognostic impact of p53 mutations or a combined larger risk group comprising AML cases with p53 mutations or LOH at 17p (with or without copy loss) on survival in AML.40,41 Together, these data provide a further refinement of survival prognostication in AML and provide a rationale for introducing array-based genomics into AML management.
Methods
Patients
Between March 2005 and November 2009, 114 patients with previously untreated AML (all non-M3) were enrolled into this study at the University of Michigan Comprehensive Cancer Center. The study was approved by the University of Michigan Institutional Review Board (IRBMED #2004-1022), and written informed consent was obtained from all patients before enrollment in accordance to the Declaration of Helsinki. Of the samples from 114 patients, 112 resulted in array-based data for paired samples (blasts vs buccal DNA [n = 107] or remission marrow DNA [n = 5]), and 2 resulted in tumor data only. The latter 2 cases were included in the outcome analysis, as they had either a genomic lesion that was also detected by karyotyping (exclusive of copy number variants [CNVs] that were identified through comparative analysis of all other cases: in one case) or lesions too large (∼ 12 Mb, ∼ 13 Mb) to be CNVs (one case).42 Cytogenetic risk stratification was determined according to Southwest Oncology Group criteria incorporating updated guidelines based on Southwest Oncology Group AML trial S0106.5
Cell isolation
Ficoll gradient separation and cryopreservation.
Peripheral blood or bone marrow mononuclear cells from AML patients were isolated as previously described.43
Microbead-based negative selection and subsequent flow cytometric sorting of leukemia specimens.
AML blast DNA used for SNP 6.0 profiling was extracted from negatively column-enriched AML cell samples as previously described43 that were further purified as follows: post-Miltenyi column samples were washed and stained with fluorescein isothiocyanate (FITC)-conjugated anti-CD33, phycoerythrin (PE)-conjugated anti-CD13, and allophycocyanin (APC)-conjugated anti-CD45 (all antibodies from eBioscience). After final washing, propidium iodide (PI) was added to a concentration of 1 μg/mL to discriminate dead cells. Sorting of cells was done on a FACSAria high-speed flow cytometer (Becton Dickinson). Live cells (ie, PI negative) were gated for blasts by identifying those cells with intermediate-intensity staining for CD45 and low- to moderate-intensity side scatter.44 CD33 and CD13 were then used to further discriminate blasts versus erythroid lineage and mature myeloid lineage cells.
Preparation of sample DNA
DNA was extracted as previously described.43
Array data analysis
The DNA was prepared for hybridization to SNP 6.0 arrays according to the manufacturer's recommendations. Affymetrix cell intensity (CEL) files for each blast and buccal sample were analyzed using Genotyping Console software Version 2.0 for initial quality control, followed by use of the Affymetrix “Birdseed” algorithm to generate tab-delimited SNP call files in text format. Call rates for the entire group of samples included in this report were between 94.93% and 99.45%, with a mean call rate of 98.33%; none of the tumor DNA samples gave out-of-bounds results.
Sample copy-number heatmap displays were obtained from CEL files through use of the freely available software, dChip (build date February 25, 2009),45 adapted to run on a 64-bit computer environment. To generate functional, practical displays of LOH, a 2-step, internally developed, Java-based software-analysis system was used. The Pre-LOH Unification Tool (PLUT) served to align all individual patient SNP calls to their respective Single Nucleotide Polymorphism Database reference SNP identification numbers and genomic physical positions before incorporation into the LOH tool version 2, an updated version of the LOH tool able to accommodate Affy SNP 6.0 array data46 (an executive copy of PLUT or of the LOH tool version 2 is available upon request).
For genomic copy-number analysis, we visually inspected parallel heatmap copy-number images of AML blast and paired normal DNA samples generated through dChipSNP and using the median smoothening functionality. Only those copy-number changes detected in blast DNA that were not found at the same position in paired normal DNA were called somatic. Using this approach, the 3 shortest identified acquired copy number (aCN) changes were 0.04, 0.068, and 0.075 Mb in length and were defined by 35, 36, and 41 consecutive SNP positions, respectively. The majority of aCN changes were defined by > 100 consecutive SNP positions (see supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
For LOH analysis between paired samples, a filter setting within the LOH tool version 2 was used, allowing visualization of individual paired SNP calls as LOH only if present within 3000 base pairs of another such call. This step filtered out many false, sporadically distributed single LOH calls due to platform noise. Further, LOH calls for at least 3 closely spaced SNPs were required to make an LOH call in any particular genomic region.
SNP 6.0 array data files for all 226 patient samples analyzed have been deposited in the Gene Expression Omnibus public database (accession no. GSE23452).
The methods for algorithmic lesion calling
Median-smoothened copy-number data were exported from dChipSNP and were transformed by raising all values to the 0.25th power to approximately stabilize the variance. Then, within each subject, the transformed normal DNA value was subtracted from the transformed tumor DNA value to create a subject-level difference summary. Using this difference summary, we then constructed running-average and -variance statistics centered on each SNP, based on uniformly weighted windows of size 30 (for the average) and 120 (for the variance). We next constructed a pseudo-Z-score as Z = √30 × A/square root (V), where A is the running average and V is the running variance. Initial lesion calls were based on the rule Z < −4 and A < −0.12 (for losses; equivalent to ∼ CN estimates of ≤ 1.3) and Z > 16 and A > 0.12 (for gains; equivalent to ∼ CN estimates of ≥ 3). These initial calls were then refined by bridging small gaps between consecutive lesion calls. Gaps between consecutive gains or losses were bridged if they were up to 1000 SNP positions in length, and the mean of the Z values within the gap was less than −2 (for a gap between consecutive losses) or greater than 2 (for a gap between consecutive gains); thus, only nominal statistical significance was required to bridge small gaps between lesions that had already been detected at high stringency thresholds. The remaining lesion calls were used for subsequent analysis without a lower size limit.
Cytogenetic analysis and fluorescence in situ hybridization (FISH)
Interphase FISH analysis for various loci was performed as previously described,47 using bacterial artificial chromosome probes as detailed in supplemental Table 2. A total of 200 interphase cells were analyzed per patient. Negative controls were prepared from cryopreserved purified AML blast cells without evidence of the respective copy-number aberrations by SNP 6.0 array profiling and were run in parallel with each hybridization.
Exon resequencing of NPM1, FLT3, p53, and CEBPA
Primers to amplify and sequence exon 12 of human nucleophosmin (nucleolar phosphoprotein B23, nutramin) (NPM1), exons 13-15 and 20 of human fms-related tyrosine kinase 3 (FLT3), exons 2-9 of human p53, and the CCAAT/enhancer binding protein (C/EBP) alpha (CEBPA) coding exon and adjacent intronic sequences were designed using the primer 3 program (http://frodo.wi.mit.edu/primer3/) Version 1.2, and sequence information was generated as previously described.43 Mutations were confirmed using paired patient buccal DNA as templates.
Statistical analysis
Overall survival was defined as the time (in days) between AML diagnosis and the patient's death. For patients still alive, the date of censoring was April 15, 2010. Uni- and bivariate analyses were based on Kaplan-Meier estimates of survivor functions. Median survival times were estimated directly from the survivor function estimates. Significance levels for groupwise comparisons in the univariate analyses assessed whether the hazard for group membership differed from 1 in a Cox proportional hazards model. These Cox models were fit using a dummy variable indicating membership in 1 of the 2 groups being compared. For bivariate analyses, Cox models were fit to subsamples defined by 1 of the 2 factors being analyzed. The reported P value assessed whether the hazard ratio for the other factor differed from 1 within the subsample.
Multivariate analyses were based on Cox proportional hazards models, with additive effects for the factors, as reported below. The reported significance levels assessed whether the hazard for a given factor differed from 1 when the other factors in the model were held fixed.
Results
Patient characteristics
Characteristics of the 114 prospectively enrolled AML patients (all previously untreated and non-M3 French-American-British subtype) analyzed in this study are summarized in Table 1 and supplemental Table 1. Sixty-eight percent, 17%, and 15% were either primary, secondary, or treatment-related AML (tAML), and 18 (16%) of the cases had p53 exon 2-9 mutations (Table 1). Eighty-five percent and 15% of patients were treated with either intensive chemotherapy up front or other types of therapy, respectively (Table 2 and supplemental Table 1). Consolidation involved a matched related or matched unrelated donor stem cell transplantation in 32% (36/114) of patients. Furthermore, 68% (78/114) of patients had died at the time of data analysis (mean and median follow-up for surviving patients was 822 and 720 days, respectively). Given that leukemia physicians occasionally select nonintensive chemotherapies for elderly high-risk patients, including patients with unfavorable cytogenetics (thus introducing a possible bias into outcome analyses), we opted to combine, for the purpose of outcome analysis, all patients into one group (all-treatment group, n = 114) and to analyze the intensively treated subgroup separately (intensive-chemo group, n = 97).
Baseline characteristics of patients
| Characteristic . | 1° AML, no. (%) . | 2° AML, no. (%) . | tAML, no. (%) . |
|---|---|---|---|
| Sample size (N = 114) | 78 (68) | 19 (17) | 17 (15) |
| Age, y | |||
| Median | 59 | 71 | 61 |
| Range | 20-80 | 50-85 | 24-73 |
| Sex | |||
| Male | 49 (63) | 12 (63) | 5 (29) |
| Female | 29 (37) | 7 (37) | 12 (71) |
| FAB classification* | |||
| M0 | 11 (14) | 2 (11) | 1 (6) |
| M1 | 10 (13) | 0 (0) | 3 (18) |
| M2 | 12 (15) | 4 (21) | 2 (12) |
| M3 | 0 (0) | 0 (0) | 0 (0) |
| M4 | 29 (37) | 5 (26) | 4 (24) |
| M5 | 8 (10) | 1 (5) | 2 (12) |
| M6 | 0 (0) | 0 (0) | 0 (0) |
| M7 | 0 (0) | 0 (0) | 0 (0) |
| Cytogenetic class | |||
| Favorable | 8 (10) | 0 (0) | 0 (0) |
| Intermediate | 44 (57) | 10 (53) | 6 (35) |
| Unfavorable | 26 (33) | 9 (47) | 11 (65) |
| Complex karyotype | |||
| Yes | 12 (15) | 6 (32) | 8 (47) |
| No | 66 (85) | 13 (68) | 9 (53) |
| 5q−/−5 cytogenetics status | |||
| Present | 10 (13) | 2 (11) | 8 (47) |
| Absent | 68 (87) | 17 (89) | 9 (53) |
| 7q−/−7 cytogenetics status | |||
| Present | 10 (13) | 4 (21) | 6 (35) |
| Absent | 68 (87) | 15 (79) | 11 (65) |
| NPM1 exon 12 status† | |||
| Mutated | 15 (19) | 1 (5) | 2 (12) |
| Wild-type | 63 (81) | 17 (89) | 15 (88) |
| p53 exons 2-9 status† | |||
| Mutated | 8 (10) | 3 (16) | 7 (41) |
| Wild-type | 69 (90) | 15 (79) | 10 (59) |
| FLT3 ITD status† | |||
| ITD present | 11 (14) | 0 (0) | 3 (18) |
| Wild-type | 67 (86) | 18 (100) | 14 (82) |
| Characteristic . | 1° AML, no. (%) . | 2° AML, no. (%) . | tAML, no. (%) . |
|---|---|---|---|
| Sample size (N = 114) | 78 (68) | 19 (17) | 17 (15) |
| Age, y | |||
| Median | 59 | 71 | 61 |
| Range | 20-80 | 50-85 | 24-73 |
| Sex | |||
| Male | 49 (63) | 12 (63) | 5 (29) |
| Female | 29 (37) | 7 (37) | 12 (71) |
| FAB classification* | |||
| M0 | 11 (14) | 2 (11) | 1 (6) |
| M1 | 10 (13) | 0 (0) | 3 (18) |
| M2 | 12 (15) | 4 (21) | 2 (12) |
| M3 | 0 (0) | 0 (0) | 0 (0) |
| M4 | 29 (37) | 5 (26) | 4 (24) |
| M5 | 8 (10) | 1 (5) | 2 (12) |
| M6 | 0 (0) | 0 (0) | 0 (0) |
| M7 | 0 (0) | 0 (0) | 0 (0) |
| Cytogenetic class | |||
| Favorable | 8 (10) | 0 (0) | 0 (0) |
| Intermediate | 44 (57) | 10 (53) | 6 (35) |
| Unfavorable | 26 (33) | 9 (47) | 11 (65) |
| Complex karyotype | |||
| Yes | 12 (15) | 6 (32) | 8 (47) |
| No | 66 (85) | 13 (68) | 9 (53) |
| 5q−/−5 cytogenetics status | |||
| Present | 10 (13) | 2 (11) | 8 (47) |
| Absent | 68 (87) | 17 (89) | 9 (53) |
| 7q−/−7 cytogenetics status | |||
| Present | 10 (13) | 4 (21) | 6 (35) |
| Absent | 68 (87) | 15 (79) | 11 (65) |
| NPM1 exon 12 status† | |||
| Mutated | 15 (19) | 1 (5) | 2 (12) |
| Wild-type | 63 (81) | 17 (89) | 15 (88) |
| p53 exons 2-9 status† | |||
| Mutated | 8 (10) | 3 (16) | 7 (41) |
| Wild-type | 69 (90) | 15 (79) | 10 (59) |
| FLT3 ITD status† | |||
| ITD present | 11 (14) | 0 (0) | 3 (18) |
| Wild-type | 67 (86) | 18 (100) | 14 (82) |
1° AML indicates de novo AML; 2° AML, AML with antecedent myelodysplasia or MPD; tAML, treatment-related AML; and ITD, internal tandem duplication.
The French-American-British (FAB) classification was unspecified in 20 samples.
Data are unavailable for NPM1 in 1 case, p53 in 2 cases, and FLT3 in 1 case.
First-line therapies for patients analyzed in this study: first-line therapies used in the AML cohort
| Characteristic . | 1° AML, no. (%) . | 2° AML, no. (%) . | tAML, no. (%) . |
|---|---|---|---|
| Sample size (N = 114) | 78 (68) | 19 (17) | 17 (15) |
| Intensive therapy (n = 97) | 72 (92) | 10 (53) | 15 (88) |
| Anthracycline + cytarabine only | 31 (40) | 5 (27) | 5 (29) |
| Anthracycline + cytarabine + gemtuzumab | 18 (23) | 0 (0) | 0 (0) |
| Anthracycline + cytarabine + cyclosporin | 13 (17) | 1 (5) | 0 (0) |
| Amonafide + cytarabine | 1 (1) | 1 (5) | 7 (41) |
| Amonafide + gemtuzumab | 1 (1) | 1 (5) | 0 (0) |
| Clofarabine | 6 (8) | 2 (11) | 1 (6) |
| Fludarabine, cytarabine, and filgrastim | 1 (1) | 0 (0) | 2 (12) |
| Gemtuzamab + zosuquidar | 1 (1) | 0 (0) | 0 (0) |
| Intensive intermediate therapy (n = 6) | 3 (4) | 2 (11) | 1 (6) |
| Azacitidine + gemtuzumab | 3 (4) | 2 (11) | 1 (6) |
| Low-dose therapy (n = 7) | 1 (1) | 6 (31) | 0 (0) |
| Decitabine | 0 (0) | 1 (5) | 0 (0) |
| Decitabine + vorinostat | 0 (0) | 2 (11) | 0 (0) |
| Lenalidomide | 0 (0) | 1 (5) | 0 (0) |
| Imatinib | 1 (1) | 0 (0) | 0 (0) |
| Investigational aurora kinase inhibitor | 0 (0) | 1 (5) | 0 (0) |
| Obatoclax mesylate | 0 (0) | 1 (5) | 0 (0) |
| Best supportive care (n = 4) | 2 (3) | 1 (5) | 1 (6) |
| Characteristic . | 1° AML, no. (%) . | 2° AML, no. (%) . | tAML, no. (%) . |
|---|---|---|---|
| Sample size (N = 114) | 78 (68) | 19 (17) | 17 (15) |
| Intensive therapy (n = 97) | 72 (92) | 10 (53) | 15 (88) |
| Anthracycline + cytarabine only | 31 (40) | 5 (27) | 5 (29) |
| Anthracycline + cytarabine + gemtuzumab | 18 (23) | 0 (0) | 0 (0) |
| Anthracycline + cytarabine + cyclosporin | 13 (17) | 1 (5) | 0 (0) |
| Amonafide + cytarabine | 1 (1) | 1 (5) | 7 (41) |
| Amonafide + gemtuzumab | 1 (1) | 1 (5) | 0 (0) |
| Clofarabine | 6 (8) | 2 (11) | 1 (6) |
| Fludarabine, cytarabine, and filgrastim | 1 (1) | 0 (0) | 2 (12) |
| Gemtuzamab + zosuquidar | 1 (1) | 0 (0) | 0 (0) |
| Intensive intermediate therapy (n = 6) | 3 (4) | 2 (11) | 1 (6) |
| Azacitidine + gemtuzumab | 3 (4) | 2 (11) | 1 (6) |
| Low-dose therapy (n = 7) | 1 (1) | 6 (31) | 0 (0) |
| Decitabine | 0 (0) | 1 (5) | 0 (0) |
| Decitabine + vorinostat | 0 (0) | 2 (11) | 0 (0) |
| Lenalidomide | 0 (0) | 1 (5) | 0 (0) |
| Imatinib | 1 (1) | 0 (0) | 0 (0) |
| Investigational aurora kinase inhibitor | 0 (0) | 1 (5) | 0 (0) |
| Obatoclax mesylate | 0 (0) | 1 (5) | 0 (0) |
| Best supportive care (n = 4) | 2 (3) | 1 (5) | 1 (6) |
1° AML indicates de novo AML; 2° AML, AML with antecedent myelodysplasia; and tAML, treatment-related AML.
Pathologic anatomy of subchromosomal genomic copy-number changes in AML defined through SNP 6.0 array profiling
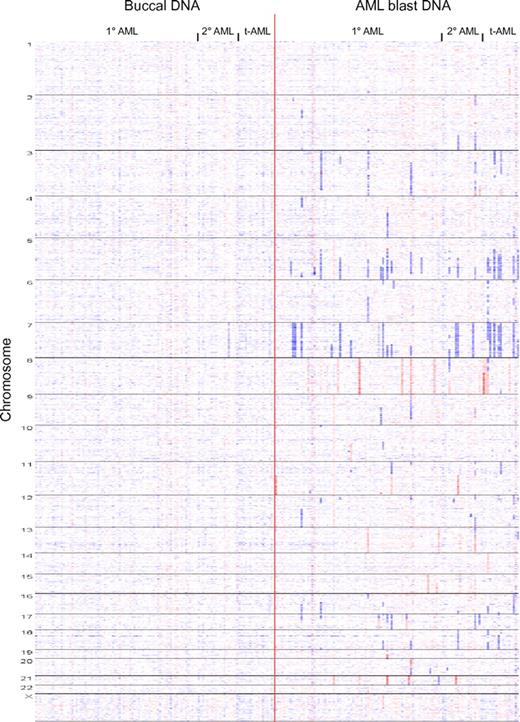
We proceeded to catalog all somatically acquired copy number changes in our AML cohort using visual inspection of simultaneous displays of dChipSNP-based copy number estimates (ie, heatmaps) for AML blast and paired buccal DNA (a whole-genome heatmap display of AML-associated aCN changes is displayed in Figure 1). LOH was catalogued in 112 cases using visual inspection of LOH displays generated using the LOHtool version 2.
Whole genome copy number heatmap display of 114 AML genomes: Copy number heatmap displays for paired DNA samples based on SNP 6.0 array profiling were generated using dChipSNP. (Left panel) Buccal DNA. (Right panel) AML blast DNA. Samples are grouped by AML subtype (primary, secondary, and treatment-related) and by chromosome number (1-22 and X). Blue indicates copy loss, red indicates copy gain. Each column represents one patient.
Whole genome copy number heatmap display of 114 AML genomes: Copy number heatmap displays for paired DNA samples based on SNP 6.0 array profiling were generated using dChipSNP. (Left panel) Buccal DNA. (Right panel) AML blast DNA. Samples are grouped by AML subtype (primary, secondary, and treatment-related) and by chromosome number (1-22 and X). Blue indicates copy loss, red indicates copy gain. Each column represents one patient.
A total of 444 somatically aCN changes were detected in 114 AML genomes (range, 0-37; see “Methods”). We detected 16 losses and 29 gains of entire chromosomes, 327 subchromosomal losses (size range, 0.04-147.12 Mb) and 72 subchromosomal gains (size range, 0.068-103.2 Mb) for a total of 399 subchromosomal aCN changes. No recurrent high-level amplifications or recurrent homozygous deletions were identified. Importantly, 21% (82/399) and 50% (199/399) of subchromosomal deletions/gains were < 1 Mb and < 5 Mb in length (see supplemental Table 1).
Acquired UPD was detected 27 times in a total of 24 AML genomes. Recurrent copy-neutral LOH (aUPD) was detected on multiple chromosomes, including chromosomes 1p, 4q, 6p, 9p, 11q, 13, 17p, and 21q (see supplemental Table 1).
Validation of SNP 6.0 array profiling results for sporadic genomic microdeletions or microgains using FISH
To provide additional validation of our datasets, we randomly selected 12 short sporadic genomic lesions (0.212-2.258 Mb in length; 9 lesions < 1 Mb in length; heatmap aCN displays are shown in supplemental Figure 1) and used FISH to confirm the presence of the aberrations. As can be seen in supplemental Table 2, all 12 lesions (and an additional 44 larger lesions) were confirmed, providing high confidence in our analytical conditions.
Internal validation of the AML study cohort using established prognostic factors
To validate our study cohort for the array-based genomic/clinical outcome analysis described below, we initially conducted univariate analyses of the most critical factors known to affect the survival of AML patients. These factors included patient age (< 60 versus ≥ 60 years), cytogenetics based on karyotyping results (grouped by risk category as unfavorable, intermediate, and favorable), NPM1 mutation status, and p53 exon 2-9 status and FLT3-ITD status; biallelic CEPBA mutations were identified in only 3 patients in this cohort). Data are summarized in supplemental Table 3 and supplemental Figure 2 (K-M plots).
Results of univariate outcome analyses of all SNP 6.0 array-detectable genomic lesions versus overall survival in AML
We initially determined the prognostic value of all SNP 6.0 array-detectable genomic aberrations (combined total of subchromosomal losses, gains, aUPD, monosomies, and trisomies) on overall survival in AML using univariate analysis.
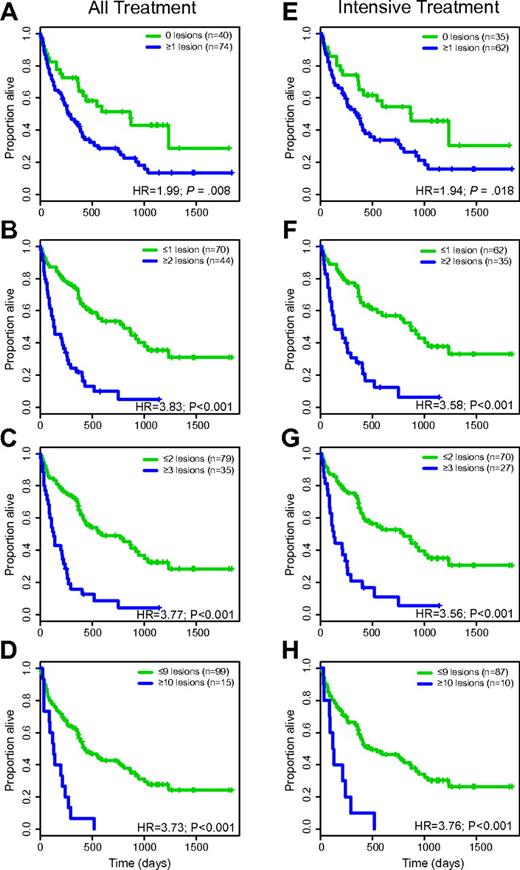
For all patients (all-treatment group), the median survival was 826 versus 264 days (hazard ratio [HR] = 1.99; P = .008) for patients with 0 (n = 40) versus ≥ 1 (n = 74) genomic lesions; 795 versus 133 days (HR = 3.83; P < .001) for patients with ≤ 1 (n = 70) versus 2 or more (n = 44) genomic lesions; 584 versus 122 days (HR = 3.77; P < .001) for patients with ≤ 2 (n = 79) versus 3 or more (n = 35) genomic lesions; and 421 versus 122 days (HR = 3.73; P < .001) for patients with ≤ 9 (n = 99) versus 10 or more (n = 15) genomic lesions, respectively. Data are summarized in Table 3, and Kaplan-Meier plots for these analyses are displayed in Figure 2A-D.
SNP 6.0 array-based lesion detection (all lesions) and survival in AML (univariate analysis)
| SNP 6.0 array genomic lesions or other risk factors . | All-treatment group . | Intensive-treatment group . | ||||||
|---|---|---|---|---|---|---|---|---|
| Survival (median, d) . | Hazard ratio . | CI . | P . | Survival (median, d) . | Hazard ratio . | CI . | P . | |
| 0 vs ≥ 1 | 826/264 | 1.99 | 1.19-3.32 | < .008 | 870/338 | 1.94 | 1.11-3.39 | .02 |
| ≤ 1 vs ≥ 2 | 795/133 | 3.83 | 2.35-6.23 | < .001 | 870/134 | 3.58 | 2.1-6.12 | < .001 |
| ≤ 2 vs ≥ 3 | 584/122 | 3.77 | 2.31-6.16 | < .001 | 795/122 | 3.56 | 2.07-6.14 | < .001 |
| ≤ 9 vs ≥ 10 | 421/122 | 3.73 | 2.03-6.86 | < .001 | 472/109 | 3.76 | 1.83-7.75 | < .001 |
| 5q−/−5 vs not 5q−/−5 | 128/472 | 3.62 | 2.09-6.26 | < .001 | 128/584 | 3.46 | 1.85-6.48 | < .001 |
| 7q−/−7 vs not 7q−/−7 | 115/433 | 2.79 | 1.62-4.79 | < .001 | 102/530 | 3.07 | 1.68-5.62 | < .001 |
| p53 exons 2-9 mutations vs not | 104/433 | 4.16 | 2.33-7.41 | < .001 | 112/520 | 4.11 | 2.12-7.99 | < .001 |
| p53 exons 2-9 mutations or 17p-LOH vs not | 122/472 | 3.84 | 2.23-6.61 | < .001 | 123/568 | 3.46 | 1.85-6.49 | < .001 |
| SNP 6.0 array genomic lesions or other risk factors . | All-treatment group . | Intensive-treatment group . | ||||||
|---|---|---|---|---|---|---|---|---|
| Survival (median, d) . | Hazard ratio . | CI . | P . | Survival (median, d) . | Hazard ratio . | CI . | P . | |
| 0 vs ≥ 1 | 826/264 | 1.99 | 1.19-3.32 | < .008 | 870/338 | 1.94 | 1.11-3.39 | .02 |
| ≤ 1 vs ≥ 2 | 795/133 | 3.83 | 2.35-6.23 | < .001 | 870/134 | 3.58 | 2.1-6.12 | < .001 |
| ≤ 2 vs ≥ 3 | 584/122 | 3.77 | 2.31-6.16 | < .001 | 795/122 | 3.56 | 2.07-6.14 | < .001 |
| ≤ 9 vs ≥ 10 | 421/122 | 3.73 | 2.03-6.86 | < .001 | 472/109 | 3.76 | 1.83-7.75 | < .001 |
| 5q−/−5 vs not 5q−/−5 | 128/472 | 3.62 | 2.09-6.26 | < .001 | 128/584 | 3.46 | 1.85-6.48 | < .001 |
| 7q−/−7 vs not 7q−/−7 | 115/433 | 2.79 | 1.62-4.79 | < .001 | 102/530 | 3.07 | 1.68-5.62 | < .001 |
| p53 exons 2-9 mutations vs not | 104/433 | 4.16 | 2.33-7.41 | < .001 | 112/520 | 4.11 | 2.12-7.99 | < .001 |
| p53 exons 2-9 mutations or 17p-LOH vs not | 122/472 | 3.84 | 2.23-6.61 | < .001 | 123/568 | 3.46 | 1.85-6.49 | < .001 |
SNP 6.0 array-based lesion cutoffs and OS in AML (Kaplan-Meier plots). (A-D) All-treatment group. (E-H) Intensive-treatment group.
SNP 6.0 array-based lesion cutoffs and OS in AML (Kaplan-Meier plots). (A-D) All-treatment group. (E-H) Intensive-treatment group.
A parallel analysis for only intensively treated patients gave the following results: the median survival was 870 versus 338 days (HR = 1.94; P = .02) for patients with 0 (n = 35) versus ≥ 1 (n = 62) genomic lesions; 870 versus 134 days (HR = 3.58; P < .001) for patients with ≤ 1 (n = 62) versus 2 or more (n = 35) genomic lesions; 795 versus 122 days (HR = 3.56; P < .001) for patients with ≤ 2 (n = 70) versus 3 or more (n = 27) genomic lesions; and 472 versus 109 days (HR = 3.76; P < .001) for patients with ≤ 9 (n = 87) versus 10 or more (n = 10) genomic lesions, respectively. Data are summarized in Table 3, and Kaplan-Meier plots for these analyses are displayed in Figure 2E-H.
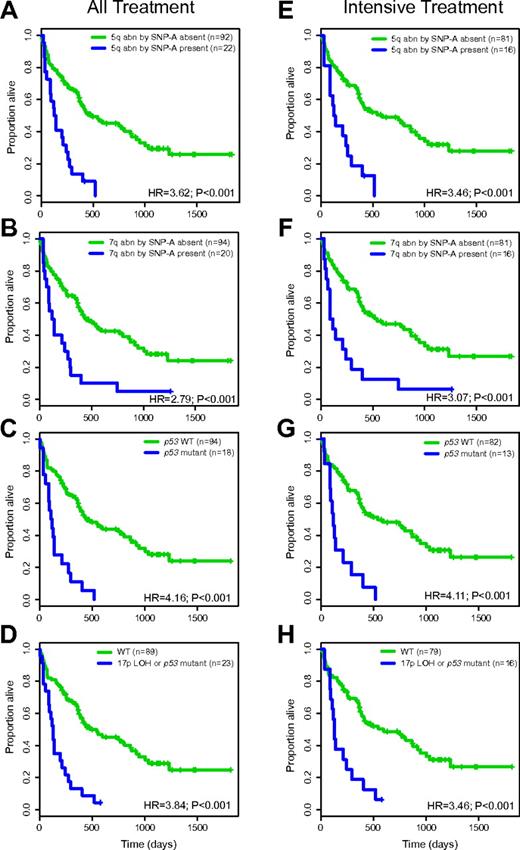
Abnormalities on 5q and 7q based on conventional karyotyping are associated with poor prognosis in AML. In this cohort, most deletions on 5q or 7q were large and readily detectable by karyotyping. Specifically, 22 patients had 5q/−5 abnormalities by SNP 6.0 array-based profiling, and 20 of these 22 patients also had known 5q/−5 abnormalities by karyotyping (FISH confirmed the SNP-A findings). Twenty patients had 7q/−7 abnormalities by SNP 6.0 array-based profiling, and 19 of these 20 also had known 7q/−7 abnormalities by karyotyping (FISH confirmed the SNP-A findings). The survival for the all-treatment group with (n = 22) or without (n = 92) 5q/−5 by SNP 6.0 array-based profiling was 128 versus 472 days (HR = 3.62; P < .001), respectively; the corresponding numbers for 7q/−7 (present n = 20; not present n = 94) were 115 versus 433 days (HR = 2.79; P = .001), respectively. The survival for intensively treated patients with (n = 16) or without (n = 81) 5q/−5 by SNP 6.0 array-based profiling was 128 versus 584 days (HR = 3.46; P < .001), respectively; the corresponding numbers for 7q/−7 (present n = 16; not present n = 81) were 102 versus 530 days (HR = 3.07; P = .001), respectively. Kaplan-Meier plots for these analyses are displayed in Figure 3A-B,E-F.
Specific SNP 6.0 array-based genomic lesions and OS in AML (Kaplan-Meier plots). (A-D) All-treatment group. (E-H) Intensive-treatment group.
Specific SNP 6.0 array-based genomic lesions and OS in AML (Kaplan-Meier plots). (A-D) All-treatment group. (E-H) Intensive-treatment group.
As part of our SNP 6.0 array-based analysis of acquired copy-number alterations, compared with LOH in AML, we detected a high frequency of copy-neutral LOH at 17p spanning p53 and an imperfect correlation between 17p alterations (aUPD-17p or 17p-copy loss) and p53 exon 2-9 mutations. In total, 23 cases had either p53 mutations or 17p-LOH or both; specifically, 5 cases had isolated p53 mutations without 17p-LOH, 5 cases had p53 mutations and deletion 17p, 8 cases had p53 mutations and aUPD at 17p, and 5 cases had wt p53 exon 2-9 and 17p-LOH (of which 2 cases did not express p53 mRNA, 2 cases did express p53 mRNA, and 1 case lacked such information). Thus, of 23 cases with 17p-LOH, 20 were p53 aberrant (null), 2 expressed wild-type p53, and 1 lacked detailed information.
Survival for the all-treatment group with (n = 18) and without (n = 94) p53 mutations was 104 and 433 days (HR = 4.16; P < .001), respectively; the corresponding numbers for the intensively treated group (present n = 13; not present n = 82) were 112 and 520 days (HR = 4.11; P = .001), respectively (within the subgroups of de novo AML and t-AML, p53 mutations were strongly negative for overall survival [OS]; HRs of 4.16 and 4.54, P < .01, respectively). Survival for the all-treatment group with (n = 23) and without (n = 89) either 17p-LOH or p53 mutations was 116 and 472 days (HR = 3.84; P < .001), respectively; the corresponding numbers for the intensively treated group (present n = 16; not present n = 79) were 122 and 520 days (HR = 3.46; P = .001), respectively. Kaplan-Meier plots for these analyses are displayed in Figure 3C-D and 3G-H.
Results of univariate outcome analyses of SNP 6.0 array-detectable genomic lesions not detectable through karyotyping versus OS in AML
To further define the individual roles served by SNP 6.0 array-based lesion detection versus conventional karyotyping in AML survival-risk prognostication, we compared lesions identified by both methods side by side. Importantly, karyotyping uniquely detected balanced chromosomal translocations, inversions, isochromosomes, marker chromosomes, and complex structural rearrangements, while SNP-array profiling uniquely detected many previously unknown small deletions and gains, as well as many hyperfragmented chromosomal regions (comprising many distinct lesions with interspersed retained chromosomal material) that appeared to be either contiguous lesions by karyotyping or where identified in patients with translocations. Both methods appeared highly complementary, in that 49% (56/114) of cases carried at least one lesion uniquely detectable through SNP 6.0 profiling, while 52% (59/114) carried at least one lesion only detected through conventional karyotyping. Approximately 347/399 = 87% of distinct lesions, as identified by SNP arrays, were not detected by karyotyping (inclusive of all hyperfragmented lesions).
We proceeded to determine the prognostic value of genomic aberrations uniquely detectable through SNP 6.0 array profiling on overall survival in AML using univariate analysis. Data are summarized in Table 4, demonstrating that the presence of such lesions alone confers substantial risk to the survival of AML patients (see below for multivariate data).
SNP 6.0 array-based uniquely detectable lesions and survival in AML (univariate analysis)
| Genomic lesions only detected by SNP 6.0 array profiling . | Survival (median, d) . | Hazard ratio . | CI . | P . |
|---|---|---|---|---|
| All-treatment group | ||||
| 0 vs ≥ 1 | 568/250 | 2.08 | 1.3-3.32 | .002 |
| ≤ 1 vs ≥ 2 | 744/133 | 3.36 | 2.08-5.44 | < .001 |
| ≤ 2 vs ≥ 3 | 520/122 | 4.31 | 2.47-7.5 | < .001 |
| ≤ 3 vs ≥ 3 | 472/122 | 3.88 | 2.2-6.83 | < .001 |
| ≤ 4 vs ≥ 4 | 433/104 | 4.31 | 2.39-7.8 | < .001 |
| ≤ 5 vs ≥ 5 | 433/104 | 4.31 | 2.39-7.8 | < .001 |
| ≤ 9 vs ≥ 10 | 421/109 | 3.86 | 2.07-7.21 | < .001 |
| Intensive-treatment group | ||||
| 0 vs ≥ 1 | 826/284 | 1.92 | 1.15-3.21 | .01 |
| ≤ 1 vs ≥ 2 | 795/169 | 3.01 | 1.76-5.14 | < .001 |
| ≤ 2 vs ≥ 3 | 520/122 | 3.85 | 1.99-7.43 | < .001 |
| ≤ 3 vs ≥ 3 | 520/109 | 3.7 | 1.89-7.25 | < .001 |
| ≤ 4 vs ≥ 4 | 520/104 | 4.11 | 2.04-8.28 | < .001 |
| ≤ 5 vs ≥ 5 | 520/104 | 4.11 | 2.04-8.28 | < .001 |
| ≤ 9 vs ≥ 10 | 472/109 | 3.76 | 1.83-7.75 | < .001 |
| Genomic lesions only detected by SNP 6.0 array profiling . | Survival (median, d) . | Hazard ratio . | CI . | P . |
|---|---|---|---|---|
| All-treatment group | ||||
| 0 vs ≥ 1 | 568/250 | 2.08 | 1.3-3.32 | .002 |
| ≤ 1 vs ≥ 2 | 744/133 | 3.36 | 2.08-5.44 | < .001 |
| ≤ 2 vs ≥ 3 | 520/122 | 4.31 | 2.47-7.5 | < .001 |
| ≤ 3 vs ≥ 3 | 472/122 | 3.88 | 2.2-6.83 | < .001 |
| ≤ 4 vs ≥ 4 | 433/104 | 4.31 | 2.39-7.8 | < .001 |
| ≤ 5 vs ≥ 5 | 433/104 | 4.31 | 2.39-7.8 | < .001 |
| ≤ 9 vs ≥ 10 | 421/109 | 3.86 | 2.07-7.21 | < .001 |
| Intensive-treatment group | ||||
| 0 vs ≥ 1 | 826/284 | 1.92 | 1.15-3.21 | .01 |
| ≤ 1 vs ≥ 2 | 795/169 | 3.01 | 1.76-5.14 | < .001 |
| ≤ 2 vs ≥ 3 | 520/122 | 3.85 | 1.99-7.43 | < .001 |
| ≤ 3 vs ≥ 3 | 520/109 | 3.7 | 1.89-7.25 | < .001 |
| ≤ 4 vs ≥ 4 | 520/104 | 4.11 | 2.04-8.28 | < .001 |
| ≤ 5 vs ≥ 5 | 520/104 | 4.11 | 2.04-8.28 | < .001 |
| ≤ 9 vs ≥ 10 | 472/109 | 3.76 | 1.83-7.75 | < .001 |
An elevated genomic lesion load as detected through SNP 6.0 array profiling predicts for low complete response (CR) rates
The response to initial treatment is of clinical importance to AML outcome. Therefore, we determined the fraction of patients that achieved CR/CRi that had SNP 6.0 profiling-based genomic lesion scores below or above defined thresholds (multiple thresholds were tested) and found that an elevated lesion load predicted for low CR/CRi rates at high significance levels, an effect mostly driven by cases with high genomic complexity. Data are summarized in Table 5.
SNP 6.0 array-based lesion thresholds and associated CR/CRi rates in AML
| SNP 6.0 array lesion threshold . | Proportion of CR/CRi in low lesion group . | Proportion of CR/CRi in high lesion group . | Z-score for log-odds ratio . | P . |
|---|---|---|---|---|
| All treatment | ||||
| 0 vs ≥ 1 | 0.8 | 0.507 | 2.95 | .003 |
| ≤ 1 vs ≥ 2 | 0.729 | 0.415 | 3.21 | .001 |
| ≤ 2 vs ≥ 3 | 0.722 | 0.344 | 3.56 | < .001 |
| ≤ 3 vs ≥ 4 | 0.708 | 0.227 | 3.77 | < .001 |
| ≤ 4 vs ≥ 5 | 0.703 | 0.2 | 3.72 | < .001 |
| Intensive chemo only | ||||
| 0 vs ≥ 1 | 0.857 | 0.574 | 2.73 | .006 |
| ≤ 1 vs ≥ 2 | 0.774 | 0.5 | 2.72 | .007 |
| ≤ 2 vs ≥ 3 | 0.771 | 0.423 | 3.19 | .002 |
| ≤ 3 vs ≥ 4 | 0.759 | 0.294 | 3.41 | .001 |
| ≤ 4 vs ≥ 5 | 0.753 | 0.267 | 3.93 | .001 |
| SNP 6.0 array lesion threshold . | Proportion of CR/CRi in low lesion group . | Proportion of CR/CRi in high lesion group . | Z-score for log-odds ratio . | P . |
|---|---|---|---|---|
| All treatment | ||||
| 0 vs ≥ 1 | 0.8 | 0.507 | 2.95 | .003 |
| ≤ 1 vs ≥ 2 | 0.729 | 0.415 | 3.21 | .001 |
| ≤ 2 vs ≥ 3 | 0.722 | 0.344 | 3.56 | < .001 |
| ≤ 3 vs ≥ 4 | 0.708 | 0.227 | 3.77 | < .001 |
| ≤ 4 vs ≥ 5 | 0.703 | 0.2 | 3.72 | < .001 |
| Intensive chemo only | ||||
| 0 vs ≥ 1 | 0.857 | 0.574 | 2.73 | .006 |
| ≤ 1 vs ≥ 2 | 0.774 | 0.5 | 2.72 | .007 |
| ≤ 2 vs ≥ 3 | 0.771 | 0.423 | 3.19 | .002 |
| ≤ 3 vs ≥ 4 | 0.759 | 0.294 | 3.41 | .001 |
| ≤ 4 vs ≥ 5 | 0.753 | 0.267 | 3.93 | .001 |
Results of bivariate outcome analyses of traditional prognostic factors and SNP 6.0 array-detectable genomic lesions versus overall survival in AML
Next we evaluated the prognostic value of SNP 6.0 array profiling-based lesion detection in the setting of either age or cytogenetics. Data are summarized in supplemental Table 4. Importantly, the detection of ≥ 2 lesions by SNP 6.0 arrays (and higher cutoffs) added negative prognostic value to both age and cytogenetics information. For example, the median survival estimates for the all-treatment group analysis of age < 60 and a SNP 6.0 array profiling-based lesion cutoff of ≤ 1 (n = 38) or ≥ 2 lesions (n = 14) were > 1200 and 182 days (HR = 5.9; P = < .001), respectively; survival estimates were 406 and 109 days for age ≥ 60 and either ≤ 1 (n = 32) or ≥ 2 lesions (n = 30; HR = 2.27; P = .004), respectively. Kaplan-Meier curves for the various SNP 6.0 array profiling-based lesion cutoffs are displayed in supplemental Figure 3A-H.
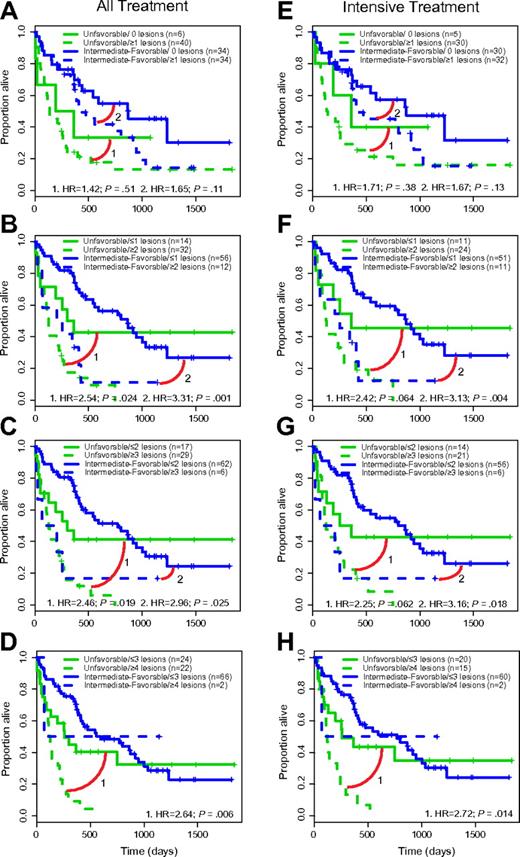
In the bivariate analyses that included the 2 genomic variables cytogenetic risk group and SNP 6.0 array profiling-based lesion count, the median survival estimates for the all-treatment group analysis of unfavorable cytogenetics and a SNP 6.0 lesion cutoff of ≤ 1 (n = 14) versus ≥ 2 lesions (n = 32) were 308 and 123 days (HR = 2.54; P = .024), respectively; survival estimates were 832 and 161 days for intermediate/favorable cytogenetics and either ≤ 1 (n = 56) or ≥ 2 lesions (n = 12; HR = 3.31; P = .001), respectively. Kaplan-Meier curves for the various SNP 6.0 array profiling-based, lesion-based cutoffs are displayed in Figure 4A-H (the data for bivariate analyses using either age or cytogenetic risk group and genomic lesions uniquely detectable through SNP 6.0 arrays are tabulated in supplemental Table 5).
SNP 6.0 array-based lesion cutoffs and cytogenetic risk group and OS in AML (Kaplan-Meier plots). (A-D) All-treatment group. (E-H) Intensive-treatment group.
SNP 6.0 array-based lesion cutoffs and cytogenetic risk group and OS in AML (Kaplan-Meier plots). (A-D) All-treatment group. (E-H) Intensive-treatment group.
Next, we analyzed the effect of p53 aberrations on survival in AML. Importantly, despite a known enrichment of p53 mutations in AML with complex karyotypes,19,20 a bivariate analysis for patients in the all-treatment group demonstrated a negative prognostic effect of p53 mutations or, alternatively, the combined variable of either p53 mutations or 17p-LOH on survival in the setting of unfavorable cytogenetics. The median survival was 104 versus 232 days (HR = 2.21; P = .02) for patients with unfavorable cytogenetics and with either p53 mutations (n = 18) or wild-type p53 (n = 27), respectively, and 122 versus 250 days for patients with unfavorable cytogenetics and with either p53 mutations or 17p-LOH present (n = 23) or absent (n = 22), respectively (HR = 2.11; P = .03). Data are displayed in supplemental Figure 4A-B.
Furthermore, the detection of a 5q/−5 abnormality by SNP 6.0 arrays demonstrated a negative prognostic effect (HR of ∼ 2; P = .04) on survival in the patients with unfavorable cytogenetics (supplemental Figure 4C,F). The detection of additional genomic lesions in the NPM1-mutated group of AML was not prognostic, although case numbers may have been too small to detect real differences (median survival of 702 and 764 days; P = .45), respectively, for NPM1 mutant cases without (n = 11) and with (n = 7) ≥ 1 SNP 6.0 array profiling-based lesions.37 Equally, the detection of genomic lesions by SNP 6.0 genomic profiling in the AML group with normal karyotype was not prognostic (median survival of 496 and 407 days; P = .28), respectively, for cases without (n = 25) and with (n = 15) ≥ 1 lesions.
Results of multivariate outcome analyses of traditional prognostic factors and SNP 6.0 array-detectable genomic lesions versus OS in AML
We proceeded with multivariate analysis, always incorporating the base variables of age- and cytogenetic-based risk category, and adding one additional variable to parallel models.
For the analysis using the all-treatment group, SNP 6.0 array profiling-based lesion detection added independent information to the negative prognostic value of age and unfavorable karyotype (HRs of 2.26 for a cutoff of ≥ 2 lesions, P < .01; HR of 2.07 for ≥ 3 lesions, P = .02; and HR of 2.28 for ≥ 4 lesions, P < .01). Results for the corresponding analysis of the intensive-treatment group were: HR of 2.1 for a cutoff of ≥ 2 lesions (P = .03); HR of 1.96 for ≥ 3 lesions (P = .06); and HR of 2.37 for ≥ 4 lesions (P = .01). Data are summarized in Table 6. Corresponding multivariate analysis data using only genomic lesions uniquely detectable through SNP 6.0 arrays gave similar results and are tabulated in supplemental Table 6.
Results of multivariate analysis of age, cytogenetic risk group, and SNP 6.0 array-based lesion cutoffs on overall survival in AML
| Variable . | All-treatment group (n = 114) . | Intensive-treatment group (n = 97) . | ||||
|---|---|---|---|---|---|---|
| Hazard ratio . | CI . | P . | Hazard ratio . | CI . | P . | |
| Age ≥ 60 vs < 60 | 2.60 | 1.54-4.41 | < .01 | 2.48 | 1.38-4.44 | < .01 |
| Cytogenetic risk group unfavorable vs not | 1.93 | 1.10-3.39 | .02 | 1.84 | 0.97-3.49 | .06 |
| No. of SNP array genomic lesions ≥ 2 vs < 2 | 2.26 | 1.27-4.05 | < .01 | 2.10 | 1.08-4.08 | .03 |
| Age ≥ 60 vs < 60 | 2.67 | 1.57-4.53 | < .01 | 2.58 | 1.44-4.61 | < .01 |
| Cytogenetic risk group unfavorable vs not | 1.94 | 1.06-3.55 | .03 | 1.86 | 0.94-3.7 | .07 |
| No. of SNP array genomic lesions ≥ 3 vs < 3 | 2.07 | 1.11-3.88 | .02 | 1.96 | 0.95-4.01 | .06 |
| Age ≥ 60 vs < 60 | 3.13 | 1.89-5.2 | < .01 | 3.17 | 1.84-5.47 | < .01 |
| Cytogenetic risk group unfavorable vs not | 2.27 | 1.33-3.88 | < .01 | 2.26 | 1.26-4.05 | .01 |
| No. of SNP array genomic lesions ≥ 4 vs < 4 | 2.28 | 1.28-4.06 | < .01 | 2.37 | 1.25-4.49 | .01 |
| Variable . | All-treatment group (n = 114) . | Intensive-treatment group (n = 97) . | ||||
|---|---|---|---|---|---|---|
| Hazard ratio . | CI . | P . | Hazard ratio . | CI . | P . | |
| Age ≥ 60 vs < 60 | 2.60 | 1.54-4.41 | < .01 | 2.48 | 1.38-4.44 | < .01 |
| Cytogenetic risk group unfavorable vs not | 1.93 | 1.10-3.39 | .02 | 1.84 | 0.97-3.49 | .06 |
| No. of SNP array genomic lesions ≥ 2 vs < 2 | 2.26 | 1.27-4.05 | < .01 | 2.10 | 1.08-4.08 | .03 |
| Age ≥ 60 vs < 60 | 2.67 | 1.57-4.53 | < .01 | 2.58 | 1.44-4.61 | < .01 |
| Cytogenetic risk group unfavorable vs not | 1.94 | 1.06-3.55 | .03 | 1.86 | 0.94-3.7 | .07 |
| No. of SNP array genomic lesions ≥ 3 vs < 3 | 2.07 | 1.11-3.88 | .02 | 1.96 | 0.95-4.01 | .06 |
| Age ≥ 60 vs < 60 | 3.13 | 1.89-5.2 | < .01 | 3.17 | 1.84-5.47 | < .01 |
| Cytogenetic risk group unfavorable vs not | 2.27 | 1.33-3.88 | < .01 | 2.26 | 1.26-4.05 | .01 |
| No. of SNP array genomic lesions ≥ 4 vs < 4 | 2.28 | 1.28-4.06 | < .01 | 2.37 | 1.25-4.49 | .01 |
Interestingly, the presence of either p53 exon 2-9 mutations or a combined variable of either p53 exon 2-9 mutations or 17p-LOH was independently adverse for survival (HRs of 2.75 [P < .01], and 2.72 [P < .01], respectively) in the all-treatment group. The corresponding data for the intensively treated group were as follows: p53 exon 2-9 mutations or the combined variable of either p53 exon 2-9 mutations or 17p-LOH had HRs of 3.23 (P < .01) and 2.84 (P = .01), respectively. Data are summarized in Table 7.
Results of multivariate analysis of age, cytogenetic risk group, and either p53 mutations or combined p53 mutations or 17p-LOH on overall survival in AML
| Variable . | All-treatment group . | Intensive-treatment group . | ||||
|---|---|---|---|---|---|---|
| Hazard ratio . | CI . | P . | Hazard ratio . | CI . | P . | |
| Age ≥ 60 vs < 60 | 3.11 | 1.88-5.15 | < .01 | 3.22 | 1.86-5.56 | < .01 |
| Cytogenetics risk group unfavorable vs not | 2.29 | 1.29-4.07 | < .01 | 2.15 | 1.12-4.11 | .02 |
| P53 exon 2-9 mutations present* vs not | 2.75 | 1.38-5.45 | < .01 | 3.23 | 1.44-7.24 | < .01 |
| Age ≥ 60 vs < 60 | 3.16 | 1.91-5.25 | < .01 | 3.29 | 1.9-5.7 | < .01 |
| Cytogenetics risk group unfavorable vs not | 2.09 | 1.13-3.87 | .02 | 2.07 | 1.05-4.08 | .03 |
| p53 exon 2-9 mutations or 17p LOH (with and without copy loss)† vs not | 2.72 | 1.35-5.47 | < .01 | 2.84 | 1.26-6.4 | .01 |
| Variable . | All-treatment group . | Intensive-treatment group . | ||||
|---|---|---|---|---|---|---|
| Hazard ratio . | CI . | P . | Hazard ratio . | CI . | P . | |
| Age ≥ 60 vs < 60 | 3.11 | 1.88-5.15 | < .01 | 3.22 | 1.86-5.56 | < .01 |
| Cytogenetics risk group unfavorable vs not | 2.29 | 1.29-4.07 | < .01 | 2.15 | 1.12-4.11 | .02 |
| P53 exon 2-9 mutations present* vs not | 2.75 | 1.38-5.45 | < .01 | 3.23 | 1.44-7.24 | < .01 |
| Age ≥ 60 vs < 60 | 3.16 | 1.91-5.25 | < .01 | 3.29 | 1.9-5.7 | < .01 |
| Cytogenetics risk group unfavorable vs not | 2.09 | 1.13-3.87 | .02 | 2.07 | 1.05-4.08 | .03 |
| p53 exon 2-9 mutations or 17p LOH (with and without copy loss)† vs not | 2.72 | 1.35-5.47 | < .01 | 2.84 | 1.26-6.4 | .01 |
n = 18 for all-treatment group; n = 13 for intensive-treatment group.
n = 23 for all-treatment group; n = 15 for intensive-treatment group.
Results of algorithmic lesion calling (uni- and multivariate analyses)
We proceeded with an analysis of the array data using a novel statistical algorithm specifically developed for this dataset (see Methods). The algorithm makes use of the availability of paired datasets (ie, tumor vs normal) as the major filter for CNV removal (through subtraction) and also takes into account variation in copy number estimates due to various factors, including platform noise.
Next, we derived univariate data for the algorithmic SNP 6.0 array-based lesion score at various cutoffs versus survival outcome. Data are displayed in supplemental Figure 5, demonstrating a negative effect of genomic lesions on survival. Subsequently, we proceeded with multivariate analyses as described above. Data are summarized in supplemental Table 7, confirming the independent prognostic impact of SNP 6.0-detectable lesions on survival outcome in AML.
Discussion
In this study, we used ultra-high-density SNP arrays to interrogate the genomes of highly purified leukemic blasts and paired buccal DNA from 114 AML patients for acquired chromosomal copy number changes and LOH. AML cases, as expected, carried variable lesion loads, with many (∼ 35%) not harboring any such lesions at all (even in this gene-level resolution analysis), while others carried greater than 30 subchromosomal losses, gains, or aUPD. Through subsequent assessments of the prognostic impact of SNP 6.0 array-based genomic lesion identification on survival in AML, we were able to determine that SNP array-based lesion detection adds independent prognostic information to the knowledge of age- and karyotype-based risk classifications. To our knowledge, this is the first SNP array-based genomics study that incorporates the most important risk factors in AML (ie, age and cytogenetics) into multivariate models that also includes novel SNP array-based variables.37 We focused on 2 such variables—the presence of ≥ 2 (or higher cutoffs) genomic lesions and the presence of either p53 mutations or 17p LOH (with and without genomic copy loss)—that each roughly double the risk of death, while controlling for the effects of age and karyotype. Therefore, SNP array-based genomic interrogations can improve the identification of high-risk AML in the setting of established, frequently used risk factors and thus complements these established adverse risk factors in the identification of patients with short OS.
This study is one of only a few SNP array-based analyses of tumor genomes in which paired patient-derived DNA samples (blasts vs buccal DNA) were used.36 Through comparison of blast-derived and paired buccal DNA, we were able to unequivocally identify small somatically acquired losses, gains, and aUPD with a high degree of certainty and to identify polymorphic CNVs directly for each patient. Of note, the visual inspection of copy-number heatmap displays (based on the validated software tool, dChipSNP) of paired samples, as applied here, is a conservative, very specific method of lesion detection; this was confirmed through FISH (for submicroscopic lesions) and karyotyping and FISH (for large deletions, such as 5q and 7q and others). This is because investigators with experience in SNP array-based lesion analysis can correct for array or sample artifacts in ways not always accomplished by statistical lesion detection methods.48 To complement the visual lesion analyses, we developed a novel algorithm for SNP 6.0 array-based lesion identification that is based on sliding window copy number averaging, simultaneous measurements/corrections of copy-number variability in a larger window flanking the averaging window, and further, statistical corrections for artifactual breaks in lesions as a consequence of wave-like fluctuations in background signals. Upon applying this novel algorithm to our dataset, we confirmed the independent prognostic impact of SNP 6.0 profiling-based lesions in AML survival. Future clinical applications of SNP 6.0 profiling would need concerted efforts at standardization of lesion calling, possibly based on a combination of visual and statistical lesion calling methods by trained cytogeneticists.48
The current study is based on DNA isolated from flow cytometer–sorted blasts. Future additional analyses (comparing DNA from purified and unpurified AML specimens, such as bone marrow or blood) are needed to clarify the importance of blast purification in SNP array-based genomic analysis of AML patient samples.
This analysis was based on AML patients enrolled prospectively at a single center, but treated with various treatment schemas. To minimize the effect of biases introduced through therapy selection by the treating physicians, we analyzed outcome data for 2 groups of patients: all patients and the subset of patients treated only with intensive chemotherapy regimens. Importantly, the parallel analyses provided essentially the same results and conclusions: that SNP 6.0 array-based lesion detection coincides with increased risk for short survival, and it identifies subgroups of AML patients for which current treatment approaches result in dismal outcome. Regarding potential mechanisms for the observed effects, the analysis of much larger AML cohorts would be needed to analyze, in detail, the relative contributions of genomic complexity versus individual recurrent genomic lesions versus mutations in specific genes, such as p53, in prognostication of the survival in AML.
In this cohort, we detected a high frequency of LOH at 17p that spans the p53 gene. Approximately half of this LOH was copy neutral (aUPD) and therefore undetectable through conventional karyotyping. We found that p53 mutations alone or in combination with 17p-LOH identified an AML subset with very short survival, and that knowledge of 17p-LOH or p53 mutations in AML patients added independent negative prognostic information to age and unfavorable karyotype results. These data therefore extend prior observations of a negative prognostic impact of deletion 17p in AML40,49 to larger patient populations and provide an additional rationale to measure p53 defects in AML in the clinical setting.
In summary, our data demonstrate that a more comprehensive assessment of genomic copy number aberrations, as enabled through use of ultra-high-resolution SNP arrays, adds independent prognostic information to AML outcome prognostication and thus constitutes an important research direction for future studies aimed at improving AML survival prognostication and, ultimately, clinical care.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful for services provided by the microarray core of the University of Michigan Comprehensive Cancer Center.
This work was supported by the National Institutes of Health through 1R01 CA136537-01 (to S.M.) and the Translational Research Program of the Leukemia & Lymphoma Society of America (to S.M.). This research was supported in part by the National Institutes of Health through the University of Michigan's Cancer Center Support Grant (5 P30 CA46592).
National Institutes of Health
Authorship
Contribution: P.O. and S.M. performed the laboratory research; B.P., H.E., L.K., M.T., and S.M. enrolled patients and contributed and analyzed clinical data; J.K. assisted with and advised on clinical data interpretation; K.S., S.S., and C.L. assisted with statistical analysis and software development for data analysis; D.R. and A.P. performed the cytogenetics and FISH analyses; S.M. conceived the study and supervised the work; and B.P., K.S., and S.M. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sami N. Malek, Department of Internal Medicine, Division of Hematology and Oncology, University of Michigan, 1500 East Medical Center Dr, Ann Arbor, MI 48109-0936; e-mail: smalek@med.umich.edu.