Genes are embedded in chromatin, which is composed of nucleosomes and many other proteins. The core histones of the nucleosomes are subject to extensive modifications that regulate the accessibility of genes in the nucleus.3 Most of these modifications are introduced into the N-terminal histone tails that extrude from the globular histone domains of the nucleosome core particle. Particularly instructive for the expression of genes are histone methylations. While di- and tri-methylation of histone H3 at lysine 4 favors the activation of genes, methylation of histone H3 at lysines 9 and 27, as well as methylation of histone H4 at lysine 20, is associated with silenced genes and/or heterochromatin.
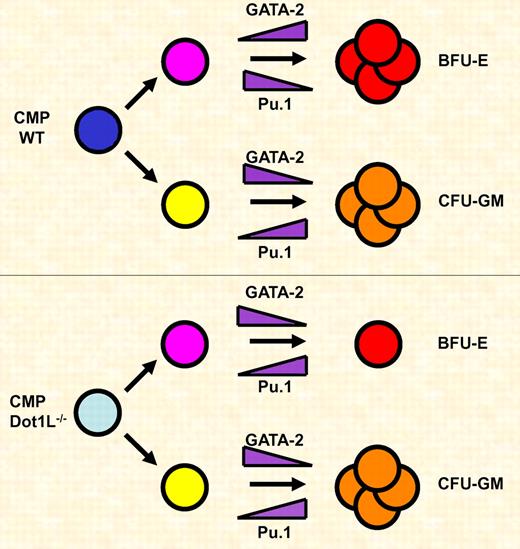
Dot1L is required for the expansion of erythroid but not myeloid progenitor cells. Common myeloid progenitor (CMP) cells differentiate along the erythroid (Bfu-E) or myeloid (CFU-GM) lineage. During erythroid differentiation GATA-2 is up-regulated and Pu.1 is down-regulated. During myeloid differentiation GATA-2 is down-regulated and Pu.1 levels increase. Dot1L deficiency causes a block in the proliferation and survival of erythroid progenitor cells, which is in part due to decreased expression of GATA-2 and increased expression of Pu.1.
Dot1L is required for the expansion of erythroid but not myeloid progenitor cells. Common myeloid progenitor (CMP) cells differentiate along the erythroid (Bfu-E) or myeloid (CFU-GM) lineage. During erythroid differentiation GATA-2 is up-regulated and Pu.1 is down-regulated. During myeloid differentiation GATA-2 is down-regulated and Pu.1 levels increase. Dot1L deficiency causes a block in the proliferation and survival of erythroid progenitor cells, which is in part due to decreased expression of GATA-2 and increased expression of Pu.1.
The histone methyltransferase Dot1L mediates the mono-, di-, or trimethylation of histone H3 at lysine residue 79 (H3K79), which is located in the globular domain of histone H3. Methylation of H3K79 is associated with actively transcribed genes suggesting a role of this modification in the process of transcription. Disruption of the yeast Dot1p gene leads to loss of telomeric silencing due to defects in the formation of heterochromatin.4 The studies in yeast suggest that Dot1-mediated H3K79 methylation restricts the binding of repressive activities to heterochromatin.
Several recent publications report on the consequences of disrupting Dot1L activity in mice.2,5,6 The studies used different experimental strategies in eliminating or strongly reducing Dot1L function, but overall the reported phenotypes are similar. The inactivation of Dot1L leads to early embryonic lethality between days 10.5 and 13.5 postcoitum, likely due to severe defects in yolk-sac angiogenesis. The total lack of H3K79 methylation demonstrates that Dot1L is the only histone methyltransferase that mediates this histone modification in mice. Furthermore, the Dot1L gene-targeting studies demonstrate that lack of H3K79 methylation causes defects in cell-cycle progression and in the stability of telomeres, which may in part be caused by reductions in the association of trimethylated H4K20 and dimethylated H3K9 with telomeres and centromeres.6
Given the complete lack of H3K79 methylation it is perhaps surprising that the defect in the Dot1L-deficient embryos is rather specific and mainly appears to affect angiogenesis and hematopoiesis, although Dot1L is expressed in many other tissues and cell types.6 However, Dot1L function may be important for the development and function of other organs and cell types at later stages during development. It will therefore be important in the future to examine mice in which Dot1L function is eliminated in a tissue and developmental stage-specific manner using conditional gene ablation strategies.
The study by Feng et al is of particular importance with respect to the function of Dot1L during hematopoiesis.2 The authors demonstrate that Dot1L deficiency leads to increased apoptosis in yolk sac–derived cells and a reduction in the formation of erythroid burst forming units (BFU-E) colonies. Interestingly, the formation of granulocyte-macrophage colony forming units (CFU-GM) is unaffected in the Dot1L-deficient embryos. Erythroid cells that do form in the Dot1L-deficient embryos do not reveal a defect in maturation, substantiated by the fact that expression of the globin genes is unperturbed. This is surprising in light of previous studies showing a correlation between H3K79 methylation and activation of the globin genes.7 It is possible that the methylation of H3K79 is more important for maintaining high-level globin gene expression in erythroid cells derived from fetal liver and bone marrow hematopoiesis, perhaps due to differences in chromatin structure in adult versus embryonic erythroid cells. Nevertheless, the results by Feng et al demonstrate that Dot1L is specifically important for the expansion and survival of embryonic erythroid progenitor cells, which in fact could cause the observed defect in yolk-sac angiogenesis.2
GATA factors play important roles in the specification of hematopoietic cell lineages.1,8,9 Previous work has shown that GATA-1 and GATA-2 control differentiation and proliferation of erythroid cells. This is in part mediated by suppression of Pu.1 gene expression in erythroid cells. On the other hand, Pu.1 is critical for the differentiation of myeloid cells and represses GATA factor function. Feng et al demonstrate that Dot1L deficiency decreases the expression of GATA-2 and increases the expression of Pu.1 in the yolk sac.2 The shift in expression levels of these 2 critical hematopoietic transcription factors likely contributes to the decrease in the number of erythroid progenitor cells observed in Dot1L-deficient embryos (see figure). It is interesting that the expression of other erythroid-specific transcription factors (eg, GATA-1 and SCL) or of components regulating signal-dependent differentiation and proliferation of erythroid cells (eg, erythropoietin receptor) is not affected by Dot1L deficiency. This further illustrates that, at least in embryonic erythroid cells, lack of H3K79 methylation does not globally affect gene expression patterns but rather inhibits expression of a selective number of genes. Perhaps it matters where a gene is located with respect to heterochromatin. If Dot1L restricts the binding of repressive activities to heterochromatin, as was shown in yeast,3 genes that are located in regions close to heterochromatin may be affected by loss of H3K79 methylation, while genes located further away may not.
In summary, the study by Feng et al reveals important functions of the histone methyltransferase Dot1L in erythropoiesis.2 The results shed light on the function of an important regulator of chromatin structure, which is also implicated in leukemogenesis.10 Future studies using conditional gene ablation will likely reveal a function of Dot1L in other cell types at later developmental stages.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■
REFERENCES
National Institutes of Health

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal