Abstract
Abstract 3387
The current study investigated the diagnostic and clinical role of genome-wide, single nucleotide polymorphism arrays (SNP-A) which can detect microscopic copy number changes in chronic myeloid leukemia (CML).
Genome-Wide Human SNP 6.0 Array (Affymetrix, CA, USA) was used for SNP-A karyotyping analysis in 132 CML patients. All patients were treated with imatinib and treatment outcomes were compared according to the presence of clonal aberrations (CAs) detected by SNP-A or additional cytogenetic abnormalities (ACAs) detected by metaphase cytogenetics (MCs).
Forty four clonal aberrations were identified in addition to t(9;22) (40 losses, 2 gains, 2 loss of heterozygosity [LOH]) that were not detected by MCs. The 9q34 deletions were found in 11% of cases, while 22q11.2 deletions were observed in 14% of cases. Nine cases harbored both 5'-ABL and 3'-BCR deletions adjacent to the t(9;22) breakpoint (7%). Copy number gains were identified at 8p and 9p, and losses at 2q, 7q, 8q, 11q, 13q, and 16p. Two cases with variant translocation showed additional deletions on the 3rd chromosome with involvement in variant translocations, which were missed by MC. We defined 3 commonly deleted regions (CDRs) on 9q34 and 22q11.2: CDR1 on 9q34 spanned approximately 162 kb between 9q34.11 and 9q34.12; CDR2 on 22q11.23 spanned 138 kb; CDR3 on 22q11.23 encompassed 102 kb.
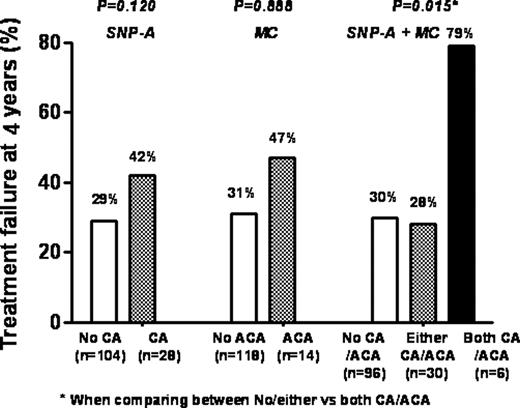
When we compared treatment outcomes according to the combination of the presence of CAs by SNP-A and/or ACAs, the patients having both CAs and ACAs showed a higher risk of failure following imatinib therapy than those with either CAs/ACAs or none (p=0.015).
The present study suggests that SNP-A analysis is a useful tool for detection of clonal aberrations in the CML genome, and could improve the prediction of prognosis in CML patients. In addition, our data suggest that SNP-A analysis is a useful tool to detect cryptic aberrations in CML genome, could improve the prediction of prognosis in CML patients and enables us to delineate CDRs on 9q34 and 22q11.2 which might be associated with CML leukemogenesis.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal