Abstract
Abstract 3227
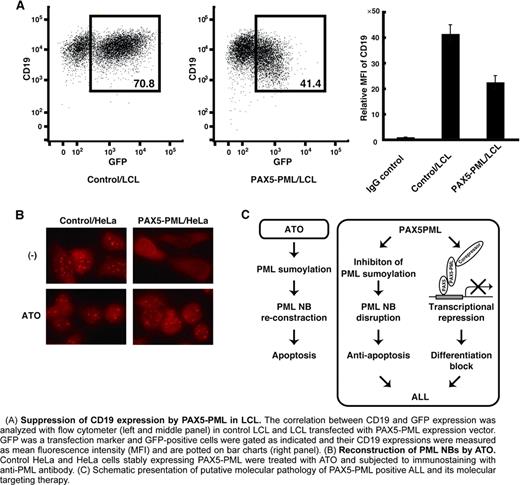
PAX5 is a transcription factor expressed in B lymphoid lineage from pro-B cell to mature B cell, and is required for B-cell development and maintenance. De-regulated and reduced PAX5 activity has been implicated in B cell malignancies both in human disease and mouse models. Furthermore, the PAX5 gene is the most frequent target of somatic mutations in childhood and adult B-progenitor acute lymphoblastic leukemia (ALL), being altered in 38.9% and 34% of cases, respectively. These mutations consist of partial or complete hemizygous deletions, homozygous deletions, partial or complete amplifications, point mutations or fusion genes. These aberrations of the PAX5 gene will impair PAX5 function more or less, and will be involved in the block of B-cell differentiation. Chromosomal translocation t(9;15)(p13;q24) was found in 2 cases of childhood ALL and resulted in an in-frame fusion of PAX5 to PML gene. PAX5 moiety of PAX5-PML retains its DNA binding domain but loses its transactivation domain suggesting PAX5-PML will be a dominant negative form of PAX5. PML is originally found as a fusion partner of RARα in PML-RARα, an oncoprotein found in acute promyeloid leukemia (APL), and is now thought to be a tumor suppressor and a pro-apoptotic factor. PML-RARα dominant-negatively affects PML function by disrupting PML nuclear bodies (NBs) where PML exerts its function and gives APL cells survival advantage. These findings give rise a speculation that PAX5-PML not only causes differentiation block by transcriptional repression of PAX5 target genes but also confers a survival advantage by inhibition of PML function. However, no functional analysis has been done for PAX5-PML. Here, we demonstrate that PAX5-PML had a dominant negative effect on both PAX5 and PML. PAX5-PML inhibited transcriptional activity of PAX5 in luciferase reporter assay. PAX5-PML expression also suppressed expression of CD19, one of the transcriptional targets of PAX5, in B-lymphoid cell line. Surprisingly, PAX5-PML hardly showed DNA binding activity in electro mobility shift assay although it retains DNA binding domain of PAX5. Further detailed analyses including, luciferase assay using PAX5-PML with DNA-binding-dead mutations, co-IP assay, and ChIP assay, suggested that transcriptional repression by PAX5-PML was independent of its DNA binding ability, that PAX5 and PAX5-PML formed a heterodimer, and that PAX5-PML bound to CD19 promoter through the association with PAX5 on the promoter. On the other hand, co-expression of PAX5-PML inhibited PML sumoylation, an essential post-translational modification for PML to form NBs, and altered PML localization from NB pattern to diffuse nuclear pattern. Furthermore, treatment with arsenic trioxide, a therapeutic reagent for APL that induces enhancement of PML sumoylation, reconstitution of PML NBs, and apoptosis in APL cells, induced recovery of PML sumoylation and reconstitution of PML NBs also in cells expressing PAX5-PML. More importantly, PAX5-PML expressing HeLa cells showed resistance to PML dependent apoptosis such as apoptosis induced by irradiation and histone deacetylace inhibitor. And arsenic trioxide treatment overcame these apoptosis resistance conferred by PAX5-PML. These data suggest the involvement of this fusion protein in the leukemogenesis of B-ALL in dual-dominant negative manner and the possibility that some cases of ALL can be treated with arsenic trioxide.
Hayakawa:Otsuka: Research Funding. Naoe:Chugai: Research Funding; Zenyaku: Research Funding; Kyowa-Kirin: Research Funding; Dainippon-Sumitomo: Research Funding; Novartis: Research Funding; Janssen: Research Funding; Otsuka: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal