Abstract
Abstract 3096
Adult T-cell leukemia/lymphoma (ATLL) is the neoplasia caused by Human T-cell leukemia virus type 1 (HTLV-1). HTLV-1-infected CD4+ T lymphocytes accumulate genetic and epigenetic changes during an extensive latency period, which can lead to neoplastic growth. Some studies have suggested that a rapidly growing clone in HTLV-1 provirus integrated T cells is selected, thus resulting in monoclonal expansion of more malignant tumor cells. In the present study, we analyzed ATLL samples using oligoarray comparative genomic hybridization (oligo-aCGH) and revealed the presence of multiple clones in the majority of ATLL patients investigated. Our data suggested that PTCL NOS is similar to ATLL with regard to polyclonal tumor cell expansion.
We performed high-resolution oligo-aCGH (Agilent Technologies Santa Clara, CA, USA) containing a 44,000 probe set against 26 paired samples comprising peripheral blood and lymph node from 13 patients with acute type ATLL. Sixteen lymph node samples from PTCL NOS patients were also examined.
All of ATLL and PTCL NOS had genomic aberrations as determined by oligo-aCGH.
1) Polyclonality of ATLL
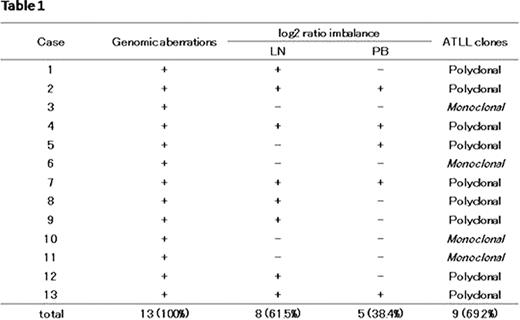
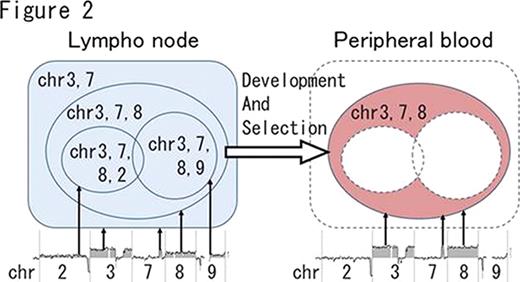
oligo-aCGH analysis showed log2 ratio average imbalances among some chromosomes in 13 of the 26 ATLL samples. For example, in the case 1 lymph node sample (Figure 1), regions of gain were detected on chromosomes 2, 3, 7, 8 and 9 as shown by the log 2 ratio in parentheses: chromosome 2 (0.10), chromosomes 3 and 7 (0.41), chromosome 8 (0.25), and chromosome 9 (0.15). These results indicate that chromosome imbalance is present in one patient sample (Figure1). Importantly, in the case 1 peripheral blood sample, regions of gain were only detected on chromosomes 3, 7 and 8, with an associated log2 ratio of almost 0.53, suggesting the absence of no chromosome imbalance. These differential genomic aberration patterns between peripheral blood and lymph node samples were found in 9 out of 13 ATLL cases (Table 1). These cases strongly indicated that the majority of ATLL patients possess polyclonal tumors.
2) Clones were selected in the peripheral blood
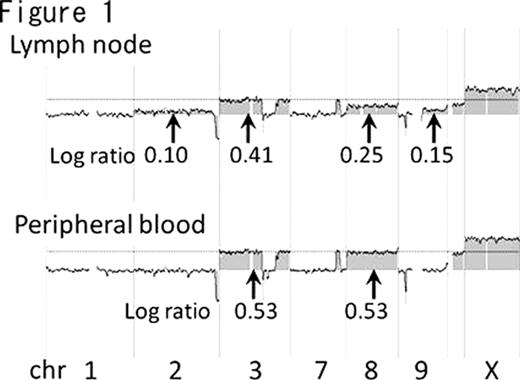
All cases examined had common genomic abnormalities between blood and lymph node samples from the same patient. However, cases 1, 2, 4, 8 and 12 indicated that selected clones from multiple clones of lymph node tumor cells had expanded in peripheral blood. A schematic representation of case 1 is shown in Figure 2.
3) Polyclonality of PTCL NOS
Eight out of 12 PTCL NOS cases showed a log2 ratio imbalance, suggesting that similar polyclonal expansion is present in PTCL NOS. An evaluation of the four remaining cases was not possible since the log2 ratio imbalance was too low.
Our results suggested that selected clones derived from a common origin may be present at the same time in acute type ATLL based on genomic analysis. The multi-clonal growth found in ATLL lymph node is also suggested to be present in PTCL NOS.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal