Abstract
Abstract 2116
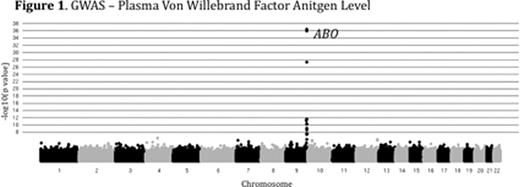
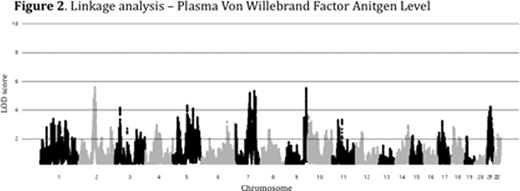
Plasma levels of von Willebrand Factor (VWF) are highly variable among healthy people and ~70% of this variability is heritable. Elevated levels of VWF are associated with thrombosis while low levels of VWF cause the common bleeding disorder, von Willebrand Disease (VWD). The genome-wide association study (GWAS) is a powerful tool for identifying common genetic variants contributing to complex traits, though less common variants are likely to be missed by this approach. We performed combined GWAS and linkage analysis in a cohort of healthy young college students. The study was approved by the University of Michigan IRB (IRBMED #2005-0080). 1189 healthy individuals (14-35 yrs of age) were recruited, comprising 507 sibships. VWF levels were determined by alphalisa assay (mean 109%, range 26 – 410%). VWF level heritability (h2) was 73% among siblings and 82% among monozygotic twins. Samples were de-identified and DNA was extracted from peripheral blood, quantified and genotyped using the Illumina Omni-1 quad SNP chip at the Broad Institute (MIT, Boston). 1182 individuals passed genotyping quality control with >97% call rates for 979,855 SNPs. Genotyping quality control was performed as part of the NIH's GENEVA program. GWAS on log transformed, age and sex adjusted VWF levels identified a major peak over the ABO locus (p<1×10-36), [Figure 1] confirming the known influence of ABO blood type on plasma VWF. Preliminary genome-wide linkage analysis (MERLIN) performed on plasma VWF levels in the 507 sibships identified 5 novel loci with peak LOD scores greater than 5 (chromosomes 2, 7, and 21), in addition to the ABO locus on chromosome 9 [Figure 2]. Other than ABO, none of these loci were detected in our GWAS analysis, or the previously reported CHARGE Consortium VWF GWAS (Smith, N et al. Circulation, 2010). Our analysis is now being extended to additional hemostasis and other related phenotypes using the same SNP data. In conclusion, linkage analysis identified novel loci contributing to the variation of VWF in healthy siblings that were undetected by GWAS. Though GWAS is an effective tool for the analysis of many complex traits, including plasma VWF variation, the majority of the heritable component of these traits remains unexplained. However, the missing heritability may be due, at least in part, to multiple rare variants at a limited number of loci. Our results suggest that genome-wide linkage analysis in multiple sibships may identify such loci and that this approach may be broadly applicable to hemostasis and other complex traits. Identification of the major loci regulating plasma VWF levels may lead to an improved prediction of bleeding and thrombotic risk and could suggest novel targets for therapeutic intervention.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal