Abstract
Abstract 2001
MCL is characterized by extreme genomic instability. Conventional techniques such as metaphase cytogenetics are unable to detect small deletions, amplifications, or uniparental disomy (UPD) in the MCL tumor genome. SNP-A analysis permits high resolution karyotyping and detection of unbalanced DNA defects, including somatic UPD. We performed SNP-A analysis on MCL tumor samples, excluded CNA present in normal controls, and assessed our results in context of clinical outcome and Ki-67 index.
With IRB approval, available frozen tissue from 18 patients diagnosed with cyclin D1-positive MCL between 1997–2006 was analyzed using high-resolution genome-wide human SNP Array 6.0 (Affymetrix). Signal intensity and SNP calls were determined using the Gene Chip Genotyping Analysis Software Version 4.0 (GTYPE) (Affymetrix). Copy number was also determined. Somatic MCL CNAs and CN-LOH were discerned from germline variants (CNV) by comparing to a database of 1535 normal controls subjected to 250K and/or SNP Array 6.0 analysis.
Clinical data was available for all patients, and 15/18 samples were subject to immunohistochemistry (IHC) for Ki67 using an automated immunostainer (Discovery; Ventana Medical Systems). Kaplan-Meier survival analysis was performed.
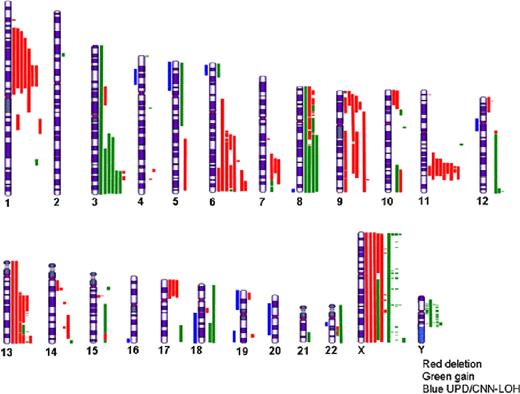
Analysis of 18 MCL patient samples revealed an average 13 CNAs (6.2 gains, 6.4 losses) and 0.4 CN-LOH per patient (Figure 1). Gains were frequently observed in 3q (33%), 8q (22%), 12q (17%), and 18q (11%). A unique 12q micro-gain in one patient narrowed the minimal common region (MCR) to linear region 130.4 – 131.3 Mbp, overlapping with an area of CN-LOH, and including candidate genes MMP17 (upregulated in invasive breast cancer), ULK1 (regulator of autophagy), and EP400 (regulator of chromatin remodeling, proliferation and apoptosis).
Recurring deletions were observed at 11q (50%), 1p, 6q (39%), 9p (33%), 13 q (28%), 9q (22%), 7q, and 17p (17%). Similar to prior studies, losses at 1p encompassing CDKN2C and FAF were seen though we further narrowed a common MCR, spanning 93.1–99.7 Mpb and including ARHGAP29 (PARG1)—previously identified in MCL by aCGH/gene expression, whose promoter is a frequent target of MCL methylation. As previously reported, losses in components of the Hippo tumor suppressor pathway were frequently affected by these recurring deletions (6q: LATS1, 9p: MOBKL2B) and by one deletion on 19p (MOBK2LA). Other high-frequency losses encompassed CDKN2A, CDKN2B, and MTAP (on 9p), RB1 and DLEU1/2/miR15a/16-1 (13q), and TP53 (17p).
Unique homozygous losses were detected at 9p (3.2-3.3 Mbp; involving only RFX3), 11q (94.5-111.7 Mbp; spanning the ATM region), and 13q (82.7-99.5 Mbp; including the miR17-92 region), and micro-deletions at 6q (121.1-121.9; GJA1/Cx43), 12p (7.5-7.6; CD163), and 13q (73.3-73.4; KLF12, and 75.2–75.3; LMO7).
CN-LOH was observed at 6p, similar to prior studies, though we found novel regions of UPD at 4p, 8q, 18q, 19q, and 22q.
An overall survival of 3.6 years and relapse-free survival of 1.3 years was observed. Survival was significantly worse among 8 pts with Ki67 >75% (OS 1.4 years, p=.003), but was unaffected by del 11q, del 9p, gain 3q, or gain 8q.
SNP-A analysis of 18 primary samples confirms that gains in 3q and 8q and losses in 11q, 6q, and 9p represent common secondary genetic lesions in MCL, and are not frequent in normal controls. We narrowed the MCR of several deletions, potential targets for gene sequencing, and confirm the presence of deletions of potential relevance to the Hippo pathway. Further analysis of our findings in light of tissue micro-array and fluorescence in-situ hybridization studies is underway, to assess pathobiologic consequences of genomic lesions as well as potential therapeutic targets.
Results of high-resolution genome-wide SNP Array of 18 MCL Patient Samples
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal