Abstract
Abstract 1473
A compact marker/suicide gene which relies on off-the-shelf reagents/pharmaceuticals would be of considerable practical utility for adoptive T-cell therapy. Marker genes enables measurement of transduction efficiency and allows sorting of transduced cells; while suicide genes allows deletion of administered T-cells in the face of toxicity. Previous marker genes include the Neomycin resistance gene, truncated nerve growth factor receptor and CD34; however these are limited by immunogenicity, unexpected biological activity and long coding region. Similarly, several suicide genes have already been described with Herpes Simplex Virus Thymidine Kinase and inducible Caspase 9 (iCasp9) in clinical use. These are limited by immunogenicity with HSVTK, limited availability of the inducing drug with iCasp9, as well as relatively long coding regions. Full-length CD20 has been tested as a suicide gene, with Rituximab as activating agent; however CD20 is biologically active when expressed in T-cells leading to spontaneous apoptosis.
We sought to generate a compact marker/suicide gene construct which enables both clinical grade sorting and effective in vivo killing using off-the-shelf clinical grade reagents/pharmaceuticals – namely CD34 cliniMACS and Rituximab. By using minimal epitopes required for CD34 cliniMACS and Rituximab binding we hoped to greatly reduce the size of the construct as well as rendering it otherwise biologically inert.
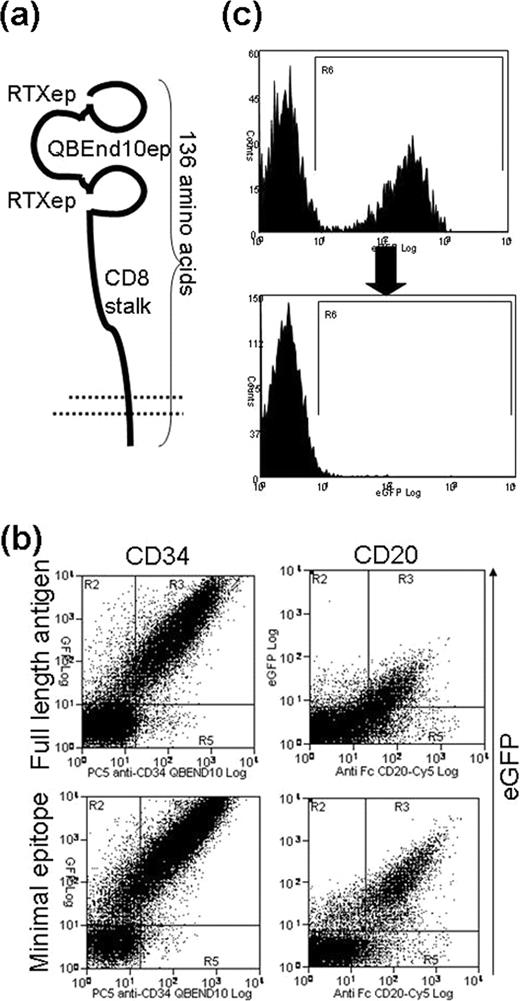
We first sought to find the minimal epitope of CD34 which binds QBEnd10, the monoclonal antibody used in Miltenyi CliniMACS CD34 selection system. Screening a library of CD34 fragments located this to the extreme amino terminus of CD34. We further minimized this to 16 residues by sequential deletion of the amino and carboxy termini. Next, we attached this epitope to the top of the CD8alpha stalk. This highly glycosylated structure acts as an effective spacer, separating the epitope from the cell membrane but is only 49 amino acids long. We found that this construct could achieve equal binding of QBEnd10 as for full-length CD34. Next, we included a minimal epitope for rituximab binding: We first tried to co-express different fragments of the CD20 major extracellular loop identified by crystallography as the rituximab binding site. These constructs failed to bind. Next, we tried linear and circular Rituximab-binding mimetopes (described by Perosa et al. J. Immunol. 2007). Inclusion of the circular mimetope (11 amino acids) afforded excellent rituximab binding. However, complement mediated killing was poor. Further variant constructs were generated: A final construct comprising of two rituximab binding epitopes flanking a single QBEnd10 epitope on the CD8 stalk [figure (a)] was allowed optimal QBend10 and rituximab binding, as well as highly effective complement mediated killing. The binding of QBEND10 to this final construct is similar to that of full-length CD34 [figure (b)]. T-cells transduced with this construct could be effectively sorted using CD34 cliniMACS. Binding of Rituximab was 3.4 fold increased relative to native CD20 [figure (b)]. Complement mediated killing could delete >97% of transduced T-cells [figure (c)]
We have created a 136 amino acid marker/suicide gene for T-cells. The translated protein is stably expressed on the cell surface after retroviral transduction. It binds QBEND10 with equal affinity to full length CD34. Further, the construct binds Rituximab, and the dual epitope design engenders highly effect complement mediated killing. Due to the small size of our construct, it can easily be co-expressed with typical T-cell engineering transgenes such as T-cell receptors or Chimeric Antigen Receptors and others allowing facile detection, cell selection as well as deletion of cells in the face of unacceptable toxicity with off the shelf clinical-grade reagents/pharmaceuticals. We are currently testing this construct in a murine model of GvHD.
(a) Cartoon showing structure of the mature marker/suicide gene. (b) Sort/suicide gene is co-expressed with eGFP. QBEND10 binding is compared with that of full-length CD34 (left); Similarly Rituximab binding to the marker/suicide construct is compared with that to full-length CD20 (right). (c) Killing efficiency after exposure to complement and rituximab gating on live cells shows deletion of practically all transduced T-cells.
(a) Cartoon showing structure of the mature marker/suicide gene. (b) Sort/suicide gene is co-expressed with eGFP. QBEND10 binding is compared with that of full-length CD34 (left); Similarly Rituximab binding to the marker/suicide construct is compared with that to full-length CD20 (right). (c) Killing efficiency after exposure to complement and rituximab gating on live cells shows deletion of practically all transduced T-cells.
Linch:Roche: Honoraria, Membership on an entity's Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal