In this issue of Blood, Devlin and colleagues use a new strategy to create a mouse model for the inherited bone marrow failure syndrome, DBA. The result, while recapitulating certain aspects of the disease and representing a positive step forward, also demonstrates that significant hurdles remain in faithfully creating a mammalian model for DBA.
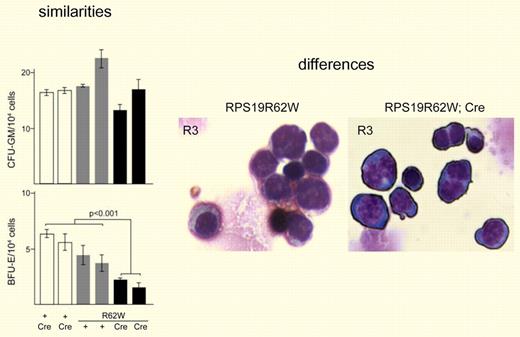
Data from Devlin et al8 that show both similarities and differences between hematologic parameters observed for RPS19R62W transgenic mice and DBA patients. Left panel: Similarities include granulocyte/macrophage precursors less affected than erythrocyte lineages. Right panel: Differences include binucleate erythroblasts not commonly found in circulation of DBA patients.
Data from Devlin et al8 that show both similarities and differences between hematologic parameters observed for RPS19R62W transgenic mice and DBA patients. Left panel: Similarities include granulocyte/macrophage precursors less affected than erythrocyte lineages. Right panel: Differences include binucleate erythroblasts not commonly found in circulation of DBA patients.
Diamond Blackfan anemia (DBA) is a rare hypoplastic anemia that typically presents within the first year of life.1 In addition to the bone marrow failure, DBA patients also manifest with a heterogeneous constellation of congenital anomalies. Nine affected genes have been identified in approximately 50% of DBA patients, all of which encode ribosomal proteins.2 Thus, DBA is the first human disease clearly established as a ribosomopathy. Although substantial progress has been made in the past few years demonstrating that haploinsufficiency for ribosomal proteins and subsequent adverse affects on ribosome assembly contribute to the underlying molecular basis of DBA, many questions remain regarding disease pathophysiology. Of particular note, the exquisite sensitivity of the developing erythron to suboptimal levels of ribosomal proteins and the timing of disease presentation in the first year of life remain unexplained.
These complex aspects of disease pathophysiology cannot be adequately assessed in cell culture models of DBA, leaving the field crying out for mammalian models. The mouse models created to date, however, have significant limitations. The majority of mutations identified in ribosomal protein genes responsible for DBA are loss-of-function mutations including complete gene deletions.3 Thus, the most straightforward mouse model for DBA would involve a gene knockout, a strategy initially used for the RPS19 gene by Matsson et al.4 Unfortunately, the hematologic profile of heterozygous mice showed no abnormalities, whereas the homozygous knockout displayed early embryonic lethality. More recently, mutations in mouse RPS19 and a second ribosomal protein gene RPS20 were identified in a forward genetic screen for dark-skinned mutants.5 In contrast to the previous attempt at a mouse model of DBA, mice harboring the dark-skinned mutations in RPS19 exhibited a mild anemia. A limited characterization of the hematologic properties of these mice revealed a relatively normal bone marrow morphology with some evidence of increased apoptosis in lin− cKit+ progenitors. Further work is necessary in the dark-skinned mouse model to determine the extent to which the underpinnings of the mild anemia observed reflect the situation in DBA patients.
Intriguingly, another mouse model that captures both some of the hematologic characteristics of DBA as well as some of its congenital anomalies lacks the FLVCR gene encoding a heme transporter present in erythroid progenitors and other cell types.6 Mutations in FLVCR have not, however, been found in DBA patients, suggesting that expression of genes like FLVCR could play a role in disease pathophysiology downstream of pathogenic mutations in ribosomal protein genes.7
In the new mouse model of DBA presented in this issue of Blood,8 Devlin et al express an RPS19 allele harboring a missense mutation (RPS19R62W) found in 4 DBA pedigrees. This mutant form of RPS19 induces hematologic phenotypes in mice with 2 normal copies of RPS19, indicating that it behaves in a dominant-negative fashion. Initial attempts to express the RPS19R62W transgene in mice resulted in early embryonic lethality. To circumvent early embryonic lethality, the authors devised a strategy to delay expression of the transgene to later stages of embryogenesis. Many of these mice die of severe anemia in utero or shortly after weaning. Survivors show a mild anemia with evidence of extramedullary compensation. Other hematopoietic lineages appear normal. Analysis of erythroid development in these mice shows a significant reduction in BFU-E and CFU-E and a 10-fold decrease in circulating reticulocytes. There was no effect on CFU-GM. Thus, these new transgenic mice recapitulate a number of hematologic parameters observed in DBA patients (see left panel of figure). And yet, there are still significant differences. Similar to other mouse models, the anemia observed in the RPS19R62W mice is mild relative to that observed in humans. The differences in anemia severity may be linked to compensating mechanisms in mice. Another distinct feature of the current mouse model are effects later in erythroid development resulting in the accumulation of binucleate erythroblasts and other evidence of improper erythroid maturation that is not observed in the DBA patient population (see right panel of figure). The binucleate cells may be a consequence of the RPS19R62W transgene inducing an arrest at the G2/M phase of the cell cycle as opposed to the G1/S arrest observed in patient cells. The basis for this difference is unknown, but it is worthwhile pointing out that the block in pre-rRNA processing observed in cells from the RPS19R62W transgenic mice is not the same as that observed in cells from patients with mutations in RPS19.9
In summary, the article by Devlin and colleagues represents a significant step toward the goal of faithfully producing a mouse model of Diamond Blackfan anemia. Moreover, the present work indicates how challenging the road has been in creating a mammalian model to study complex developmental aspects of DBA pathophysiology.
Conflict-of-interest disclosure: The authors declare no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal