Abstract
As the principal component of the membrane skeleton, spectrin confers integrity and flexibility to red cell membranes. Although this network involves many interactions, the most common hemolytic anemia mutations that disrupt erythrocyte morphology affect the spectrin tetramerization domains. Although much is known clinically about the resulting conditions (hereditary elliptocytosis and pyropoikilocytosis), the detailed structural basis for spectrin tetramerization and its disruption by hereditary anemia mutations remains elusive. Thus, to provide further insights into spectrin assembly and tetramer site mutations, a crystal structure of the spectrin tetramerization domain complex has been determined. Architecturally, this complex shows striking resemblance to multirepeat spectrin fragments, with the interacting tetramer site region forming a central, composite repeat. This structure identifies conformational changes in α-spectrin that occur upon binding to β-spectrin, and it reports the first structure of the β-spectrin tetramerization domain. Analysis of the interaction surfaces indicates an extensive interface dominated by hydrophobic contacts and supplemented by electrostatic complementarity. Analysis of evolutionarily conserved residues suggests additional surfaces that may form important interactions. Finally, mapping of hereditary anemia-related mutations onto the structure demonstrate that most, but not all, local hereditary anemia mutations map to the interacting domains. The potential molecular effects of these mutations are described.
Introduction
Preservation of red cell integrity and flexibility is conferred through an intracellular assembly of proteins known as the membrane skeleton.1-3 Through specific interactions between integral membrane proteins, adaptor proteins, and spectrin, the membrane skeleton is effectively tethered to the cell membrane. Disorders that lead to deficiencies or defects in these membrane skeleton components often disrupt the properties of this stabilizing scaffold and consequently manifest themselves clinically in a common group of disorders known as hereditary elliptocytosis (HE), hereditary pyropoikilocytosis (HPP), and hereditary spherocytosis.3-6 Although clinical, biochemical, and genetic heterogeneity add to the complexity of these syndromes, the hallmark of these disorders is the presence of deformed and fragmented erythrocytes on peripheral blood smears.3 From a molecular perspective, qualitative and quantitative disruptions of α-spectrin, β-spectrin, ankyrin, protein 4.1R, or glycophorin C all have been linked to mechanical weakness or fragility of the erythrocyte membrane skeleton. However, it is the principal component of the red cell membrane skeleton, spectrin, to which the majority of the HE and HPP disorders have been attributed.7
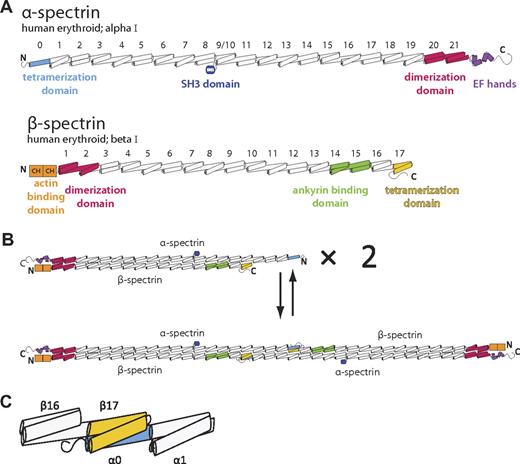
Spectrins (Figure 1A) and other members of the spectrin superfamily are characterized by multiple, tandem, spectrin-type repeats.8,9 These homologous domains are each roughly 106 amino acids in length, adopt a 3-helix bundle fold with a heptad repeat pattern of hydrophobic and hydrophilic residues, and are connected to adjacent repeats by ordered, α-helical linkers. In the case of the erythrocyte membrane skeleton, spectrin assembly begins with a lateral heterodimeric association of α and β subunits (Figure 1B top) that is nucleated by specialized dimerization repeats located near the C-terminus of α-spectrin and the N-terminus of β-spectrin.10-12 Subsequently, head-to-head interactions between 2 α/β dimers lead to the formation of a spectrin heterotetramer (Figure 1B bottom), the predominant form of the molecule in the erythroid membrane skeleton. Biochemical, biophysical, and clinical data have implicated regions near the N-terminus of α-spectrin and C-terminus of β-spectrin as being responsible for tetramer formation. Furthermore, on the basis of the length and continued heptad repeat in these regions, it had been hypothesized that these regions (α-spectrin partial repeat 0 and β-spectrin partial repeat 17) form a composite 3-helix bundle that may bear similarity to complete spectrin repeats (Figure 1C).13-15
Schematic of human erythrocyte spectrin. (A) Both the α (top) and β (bottom) isoforms of erythroid spectrin are composed of many tandem 3-helix bundle motifs (each helix is depicted as a cylinder). In the case of α-spectrin, 20 full repeats and 1 partial repeat at the N-terminus (light blue) make up the majority of the molecule. For historical reasons, the SH3 domain, located between the B and C helix of repeat α9, is assigned as the α10 motif. Similarly, β-spectrin possesses 16 full repeats and 1 partial repeat at the C-terminus (yellow). Selected regions of each spectrin isoform with a particular structure or function are colored. (B) Spectrin assembly in red cells begins with dimerization of α and β chains that is nucleated by specialized dimerization repeats (red). Subsequent tetramer formation occurs through head-to-head interaction of the tetramerization domains. (C) An enlarged schematic of the molecular components presented in this study that includes α-spectrin repeats 0-1 and β-spectrin repeats 16-17.
Schematic of human erythrocyte spectrin. (A) Both the α (top) and β (bottom) isoforms of erythroid spectrin are composed of many tandem 3-helix bundle motifs (each helix is depicted as a cylinder). In the case of α-spectrin, 20 full repeats and 1 partial repeat at the N-terminus (light blue) make up the majority of the molecule. For historical reasons, the SH3 domain, located between the B and C helix of repeat α9, is assigned as the α10 motif. Similarly, β-spectrin possesses 16 full repeats and 1 partial repeat at the C-terminus (yellow). Selected regions of each spectrin isoform with a particular structure or function are colored. (B) Spectrin assembly in red cells begins with dimerization of α and β chains that is nucleated by specialized dimerization repeats (red). Subsequent tetramer formation occurs through head-to-head interaction of the tetramerization domains. (C) An enlarged schematic of the molecular components presented in this study that includes α-spectrin repeats 0-1 and β-spectrin repeats 16-17.
Given the necessity of spectrin tetramerization for the maintenance and stability of the red cell membrane, understanding the atomic basis for these intermolecular interactions can provide insight into the nature of HE and HPP. To address this, we have crystallized and determined an atomic structure of the complex of the human erythroid tetramerization domains consisting of α-spectrin repeats 0-1 and β-spectrin repeats 16-17 to 2.8 Å resolution. This structure is the first of a biologically relevant, noncovalent complex between spectrin domains and the first to depict the β-spectrin tetramerization domain.
In addition to validating the long-standing hypothesis that the interaction between the tetramerization domains recapitulates a 3-helix bundle,16 the structural data indicate the relative positions of the partial domains with respect to one another and to their adjacent complete spectrin repeats. The overall orientation among the 3 repeats displays a striking resemblance to that of βII-spectrin repeats 14-16, thus validating that this complex truly does form a composite spectrin repeat. This likeness to complete spectrin repeats is further exemplified by the α-helical linkers that connect both partial repeats α0 and β17 to their adjacent domains. Overall, the structure is largely complete, although the first 18 amino acids of α-spectrin were disordered, as has been noted in other structures of this region, and thus were not observed. Other regions that showed disorder or variability in the structure included an interrupted helix in the C-terminal portion of β17 and the A/B loop of repeat β16.
The interface of the complex is dominated by hydrophobic contacts with limited electrostatic interactions occurring among positively charged residues of partial repeat α0 and negatively charged residues of partial repeat β17. Mapping of conserved residues onto the molecular surface revealed that not only was the interaction interface conserved but so too was one surface of the assembled complex, hinting that this domain may be involved in other biologically relevant interactions. Finally, a mapping and analysis of known mutations that cause clinical disease underscores the importance of this interaction and presents probable explanations for the molecular basis of tetramerization defects.
Methods
Cloning, expression, and purification of spectrin fragments
Constructs encompassing the tetramerization domains of human erythroid α-spectrin (repeats 0-1; residues 1-158) and human erythroid β-spectrin (repeats 16-17; residues 1902-2084) were individually cloned as glutathione-S-transferase fusion proteins into the vector pGEX-2T. The expression and purification of the individual spectrin fragments has previously been described.17 Purified α0-1 and β16-17 were mixed in a 1:1 molar ratio and incubated on ice overnight. The complex was then purified by gel filtration with the use of S-100 resin (Sigma-Aldrich) equilibrated with 10mM Tris, 0.1M NaCl, 1mM EDTA (ethylenediaminetetraacetic acid), and 1mM dithiothreitol (DTT), pH 8.0, or on a Super G3000 SWXL column (Tosoh Corporation) equilibrated in 10mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), 0.1M NaCl, and 1mM (tris(2-carboxyethyl)phosphine), pH 7.4. Once purified, the complex was concentrated to 5 to 10 mg/mL and stored at 4°C. Three cysteine mutants were constructed on the α0-1 background (R60C, A99C, and A131C) by the use of QuikChange site-directed mutagenesis (Agilent Technologies) for use in mercury derivatization. Expression of selenomethionine-substituted proteins was performed by the use of standard methods.18 Purification of mutant and selenomethionine protein was identical to that of the wild type.
Crystallization
Crystals of the α0-1/β16-17 complex were grown by hanging-drop vapor diffusion on siliconized glass coverslips (Hampton Research). Two microliters of purified protein solution (5-10 mg/mL) was mixed with an equal volume of reservoir solution containing 0.1M 2-(N-morpholino)ethanesulfonic acid, pH 6.5; 10% PEG-6000; 10% glycerol; and 5mM DTT and suspended over 0.75 mL of reservoir solution. Rectangular prismatic crystals (∼ 500 μm × 50 μm × 50 μm) grew at 4°C within 12 hours. Crystals were harvested and frozen in liquid nitrogen after serial transfer to 0.1M MES, pH 6.5; 10% PEG-6000; 10% glycerol; 5mM DTT; and 25% ethylene glycol in increasing steps of 5% ethylene glycol.
Data collection, crystal parameters, structure determination, and refinement
Data were collected at 100 K with the use of synchrotron radiation at the Advanced Photon Source Life Sciences Collaborative Access Team beamline at Argonne National Laboratory. Initial data for this project also were collected at the X29 beamline at the National Synchrotron Light Source at Brookhaven National Laboratory. Data were processed with XDS19 and scaled with SCALA.20 Additional processing was performed with programs from the CCP4 suite.20 Data for the ethyl mercury phosphate (EMP) derivative crystals (presented in the supplemental Table, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) were processed in HKL2000.21
The crystals belong to space group I41 with a = 213.9 Å b = 213.9 Å, c = 31.5 Å, α = β = γ = 90°, with one complex per asymmetric unit and with a solvent content of 72%. An experimental map was obtained by the use of data collected from EMP-derivatized crystals containing 2 separate cysteine-mutagenized constructs (α0-1 A99C and α0-1 A131C) with wild-type β16-17 at 1.01 Å and one doubly-labeled crystal containing both selenomethionine α0-1 and selenomethionine β16-17 collected at 0.979 Å (supplemental Table 1). Crystals for the purpose of derivatization were grown in the absence of reducing agents; after the crystals formed, EMP was added to the crystallization drop at a final concentration of 5mM for 16 hours before harvesting.
Multiple isomorphous replacement anomalous scattering phasing was carried out by use of the program autoSHARP,22 which correctly located the labeled cysteines and selenomethionines in the asymmetric unit. The initial solvent-flattened map was of good quality, and the 3-helix bundles of spectrin were recognized easily. An initial partial model was constructed with the low-resolution helix build module of ARP/wARP.23 Subsequent manual model building and inspection were carried out with the program Coot.24 Model refinement was performed with REFMAC5.25 Even after refinement, not all regions of the molecules were visible, particularly in the N-terminal region of the α subunit.
Proper placement of side chains was validated by use of the EMP- and selenomethionine-derivatized crystals that provided peaks in the anomalous difference map that were used as guides to register the sequence with both cysteines and methionines. TLS refinement was carried out by dividing the structure into 3 groups. The final model consists of residues: 19-158 for the α subunit (chain A) and 1900-1931 and 1934-2083 for the β subunit (chain B). The final model has an Rfactor and Rfree that converged at 26.8% and 29.9%, respectively, with a root mean square deviation (rmsd) of 0.010 Å and 1.203° for bond lengths and bond angles, respectively, and with all residues within the favored regions of the Ramachandran plot. Statistics are listed in Table 1 and supplemental Table 1.
Crystallographic statistics
| . | α0-1/β16-17 . |
|---|---|
| Data collection | |
| Detector type/source | MarCCD (Rayonix)/Advanced Photo Source |
| Wavelength, Å | 0.97872 |
| Resolution,* Å | 40.0-2.80 (2.92-2.80) |
| Measured reflections | 54 695 (6817) |
| Unique reflections | 18 015 (2195) |
| Completeness | 98.6 (99.3) |
| Multiplicity | 3.0 (3.1) |
| Mean [I/σ(I)] | 13.5 (3.4) |
| Rsym, %† | 5.7 (29.7) |
| Rmeas,%‡ | 7.0 (35.8) |
| Refinement | |
| Resolution | 35.0-2.80 (2.87-2.80) |
| Number of reflections working/test | 17 085/922 (1251/76) |
| R-work, %§ | 26.8 (40.9) |
| R-free, %‖ | 29.9 (40.5) |
| Protein atoms | 2673 |
| Bond lengths | 0.011 |
| Bond angles | 1.24 |
| . | α0-1/β16-17 . |
|---|---|
| Data collection | |
| Detector type/source | MarCCD (Rayonix)/Advanced Photo Source |
| Wavelength, Å | 0.97872 |
| Resolution,* Å | 40.0-2.80 (2.92-2.80) |
| Measured reflections | 54 695 (6817) |
| Unique reflections | 18 015 (2195) |
| Completeness | 98.6 (99.3) |
| Multiplicity | 3.0 (3.1) |
| Mean [I/σ(I)] | 13.5 (3.4) |
| Rsym, %† | 5.7 (29.7) |
| Rmeas,%‡ | 7.0 (35.8) |
| Refinement | |
| Resolution | 35.0-2.80 (2.87-2.80) |
| Number of reflections working/test | 17 085/922 (1251/76) |
| R-work, %§ | 26.8 (40.9) |
| R-free, %‖ | 29.9 (40.5) |
| Protein atoms | 2673 |
| Bond lengths | 0.011 |
| Bond angles | 1.24 |
Numbers in parentheses correspond to the highest-resolution shell throughout.
Rsym = Σ I−<I> /ΣI, where I is the observed intensity and <I> the average intensity obtained from multiple measurements.
Rmeas as described in Diederichs and Karplus.29
R-work=Σ Fo − Fc /Σ Fo, where Fo is the observed structure factor amplitude and Fc the calculated structure factor amplitude.
Rfree: R-factor determined on the basis of 5% of the data excluded from refinement.
Data deposition
Coordinates and structure factors have been deposited in the Protein Data Bank and are accessible through accession number 3LBX (http://www.rcsb.org/pdb/explore/explore.do?structureId=3LBX).
Results
Protein purification, complex formation, and characterization
A structure of the complex between the tetramerization domains of human erythroid α- and β-spectrin was determined by x-ray crystallography. Selection and design of the component protein fragments were determined on the basis of previous biochemical and biophysical studies that demonstrated human erythrocyte α0-1 (residues 1-158) and β16-17 (residues 1902-2084) were not only capable of forming a complex but were each stable and homogenous in solution.17 After purification of the individual protein fragments, the complex was reconstituted and purified by gel filtration. The complex was then characterized biophysically and used for crystallization. As expected, circular dichroism wavelength scans confirmed the large amount of α-helical secondary structure of the complex (supplemental Figure 1), consistent with previous studies.30,31 In addition, analytical ultracentrifugation to sedimentation equilibrium showed the complex to be heterodimeric in solution with a molecular weight corresponding to that of a 1:1 complex at 4°C and a dissociation constant that is temperature dependent (supplemental Figure 2).
Structure of the spectrin α0-1/β16-17 complex
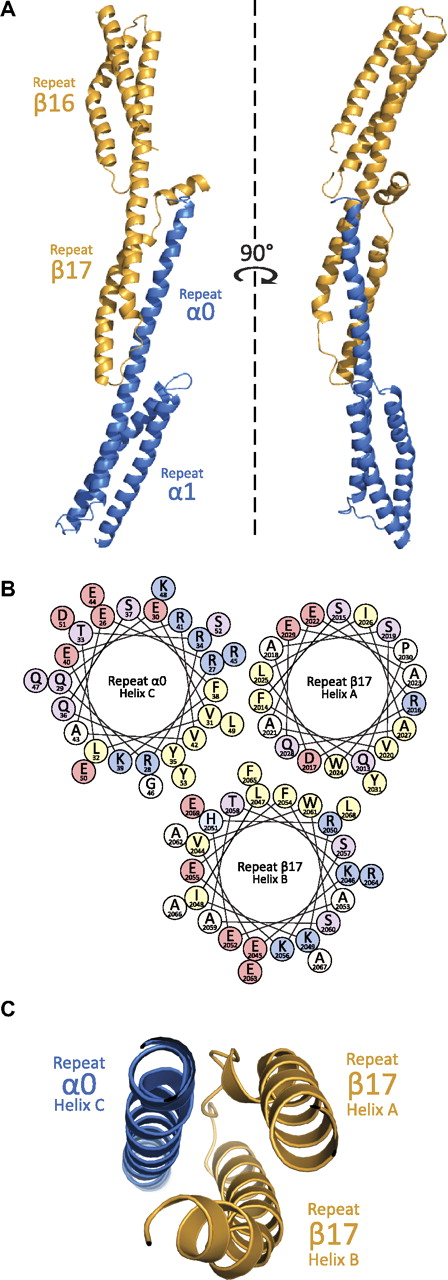
The general structure of the α0-1/β16-17 complex (Figure 2) can be divided into 3 regions: repeat 1 of α-spectrin, repeat 16 of β-spectrin, and the composite 3-helix bundle created by the partial repeats α0 and β17. Each of these regions displays a canonical spectrin fold consisting of a triple-helical bundle with hydrophobic amino acids (positions a and d in the heptad repeat) facing the interior of the bundle, and hydrophilic side chains (positions e and g) being solvent-exposed. As has been observed with all other phased, multirepeat spectrin structures to date, adjacent repeats are connected with α-helical linkers.32-38 As predicted by structure-based homology modeling and biophysical measurements, the crystal structure confirms a complex dominated by α-helices. Finally, although the aforementioned studies contributed significantly in modeling the interactions present at the tetramerization interface, it is only with the structure of this complex that the precise nature of the molecular interactions can be discerned (Figure 3).
Structural features of the assembled tetramerization site. (A) Crystallographic structure determination of the assembled tetramerization site showed a complex akin to many 3-repeat spectrin structures. As can be shown from the ribbon diagram (α-helices are depicted as coils), the interaction surface is between α-spectrin (blue) partial repeat 0 and β-spectrin (yellow) partial repeat 17. (B) The helical wheel diagram of the interacting helices shows a clustering of hydrophobic residues (yellow) at the core of the 3-helix bundle. Each helical wheel depicts the radial positions of side chains upon projection down the helical axis. The helices are roughly positioned according to their placement in the structure. Residues are labeled according to single-letter code with each being color coded on the basis of properties of the side chains (positively-charged = blue, negatively charged = pink, polar = purple, hydrophobic = yellow, other = white/beige). (C) A cut-away view of the ribbon diagram near the core of the interacting surface; the orientation of the helices is similar to that in panel B.
Structural features of the assembled tetramerization site. (A) Crystallographic structure determination of the assembled tetramerization site showed a complex akin to many 3-repeat spectrin structures. As can be shown from the ribbon diagram (α-helices are depicted as coils), the interaction surface is between α-spectrin (blue) partial repeat 0 and β-spectrin (yellow) partial repeat 17. (B) The helical wheel diagram of the interacting helices shows a clustering of hydrophobic residues (yellow) at the core of the 3-helix bundle. Each helical wheel depicts the radial positions of side chains upon projection down the helical axis. The helices are roughly positioned according to their placement in the structure. Residues are labeled according to single-letter code with each being color coded on the basis of properties of the side chains (positively-charged = blue, negatively charged = pink, polar = purple, hydrophobic = yellow, other = white/beige). (C) A cut-away view of the ribbon diagram near the core of the interacting surface; the orientation of the helices is similar to that in panel B.
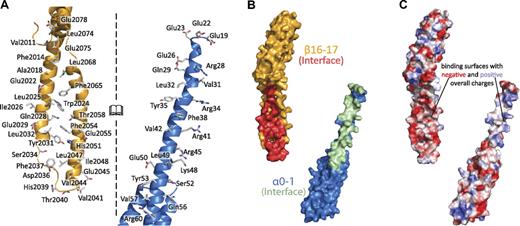
Interactions along the spectrin tetramerization domain interface are extensive. (A) The interacting surfaces of the tetramerization domain span nearly the entire composite spectrin repeat (α-spectrin shown in blue, β-spectrin shown in yellow). The interface of the complex is opened to the reader such that the interacting surface on each molecule is viewable. Specific side chains that make contact in the crystal structure are shown as sticks and labeled. (B) The molecular surfaces of α0-1 (blue, surface that interacts with β16-17 in green) and β16-17 (yellow, surface that interacts with α0-1 in red) indicate the extent of binding along these 2 molecules. (C) The electrostatic surfaces of α0 and β17 show some charge complementarity, the electrostatic map was contoured at 15 ± kBT/e. Views in panels B and C are similar to those in panel A, with both of the interacting surfaces turned to face the reader.
Interactions along the spectrin tetramerization domain interface are extensive. (A) The interacting surfaces of the tetramerization domain span nearly the entire composite spectrin repeat (α-spectrin shown in blue, β-spectrin shown in yellow). The interface of the complex is opened to the reader such that the interacting surface on each molecule is viewable. Specific side chains that make contact in the crystal structure are shown as sticks and labeled. (B) The molecular surfaces of α0-1 (blue, surface that interacts with β16-17 in green) and β16-17 (yellow, surface that interacts with α0-1 in red) indicate the extent of binding along these 2 molecules. (C) The electrostatic surfaces of α0 and β17 show some charge complementarity, the electrostatic map was contoured at 15 ± kBT/e. Views in panels B and C are similar to those in panel A, with both of the interacting surfaces turned to face the reader.
The molecular model derived from the crystallographic data are largely complete. Although the N-terminal 18 amino acids of the α subunit were present and required for crystallization, this region was not observed in the density map indicating disorder. Various predictions, as well as biochemical experiments, demonstrated that this region is not essential for tetramer formation, and its persistence as a disordered molecular fragment under crystallographic conditions is consistent with that observed in solution.39 Furthermore, although the presence of disordered regions in spectrin structures is not unusual,33,34 the molecular context of this portion of the molecule (ie, being at the terminus of the molecule) likely lends itself to disorder. Other regions of the structure that exhibited significant flexibility or disorder included the A/B loop of repeat β16 and the C-terminal tail of β17. Near the C-terminus of repeat β17, a single proline (Pro2071) residue imposes a kink that disrupts the helical fold. The most C-terminal residues (2072-2083) were observable, but significantly disordered, in the crystal structure. The density corresponding to this ending portion of the molecule had an overall helical character; however, the side chains were poorly visible, suggesting significant variability of the structure in this region (supplemental Figure 3).
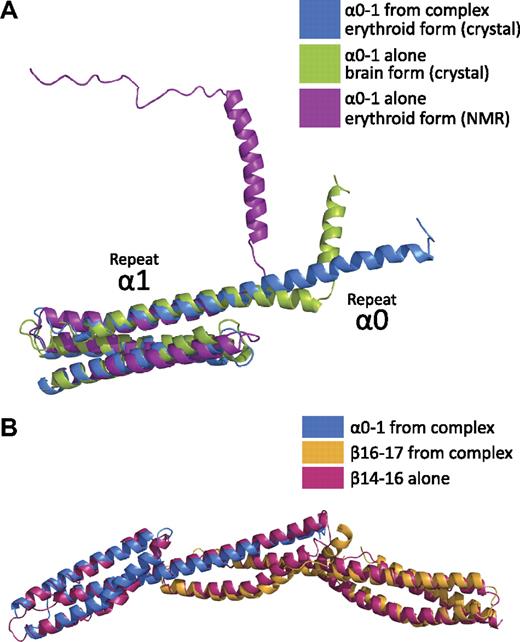
Comparison of the α0-1/β16-17 complex with other 3-repeat spectrin structures reveals a striking preservation of the overall spectrin fold in the composite repeat of α0/β17 (Figure 4).39 In addition, the relative orientations of these repeats superimposed surprisingly well with the ankyrin-binding region fragment of human brain β-spectrin repeats 14-1632 with a rmsd of 2.4 Å.
Comparison with other spectrin structures shows that the composite 3-helix bundle recapitulates a spectrin repeat. (A) Comparison of human erythroid spectrin repeats α0-1 from the crystal structure of the complex (α0-1, blue) to the structure of α0-1 in solution (purple)39 and to a crystal structure of human brain α0-1 spectrin (green)42 indicates stabilization of the helical linker between repeats 0 and 1 in the head-to-head α/β complex. In contrast to the NMR structure and crystal structure, the entirety of the repeat α0 is helical when in complex with β16-17. (B) Superposition of the structure of the tetramerization domain (blue and yellow) with the 3-repeat structure of human brain β-spectrin repeats 14-16 (red)32 demonstrate that the composite repeat highly resembles an intact spectrin repeat.
Comparison with other spectrin structures shows that the composite 3-helix bundle recapitulates a spectrin repeat. (A) Comparison of human erythroid spectrin repeats α0-1 from the crystal structure of the complex (α0-1, blue) to the structure of α0-1 in solution (purple)39 and to a crystal structure of human brain α0-1 spectrin (green)42 indicates stabilization of the helical linker between repeats 0 and 1 in the head-to-head α/β complex. In contrast to the NMR structure and crystal structure, the entirety of the repeat α0 is helical when in complex with β16-17. (B) Superposition of the structure of the tetramerization domain (blue and yellow) with the 3-repeat structure of human brain β-spectrin repeats 14-16 (red)32 demonstrate that the composite repeat highly resembles an intact spectrin repeat.
The interaction interface
On the basis of the long-standing hypothesis that the interaction surface between the partial repeats would mimic the interactions observed along the length of complete spectrin repeats, an interface rich in hydrophobic interactions was expected. Mapping of the intermolecular contacts observed in the structure (Figure 3; supplemental Figure 440 ) demonstrated that the binding surface is indeed dominated by hydrophobic contacts. In fact, relatively few electrostatic interactions were observed: Arg28alpha with Glu2022beta, Arg34alpha with Glu2022beta, and Glu2029beta. Moreover, there was only 1 salt bridge (between Arg41alpha and Glu2029beta) and only a few interchain hydrogen bonds (Gln29alpha with Leu2068beta, Arg45alpha with Asp2036beta, and Ser52alpha with Phe2037beta). Interestingly, the crystal structure revealed a much larger number (n = 53) of interactions than described previously by homology modeling (n = 15).41 Approximately one-half of the predicted 15 interactions were verified, and the rmsd difference for main chain atoms between the models was 5.0 Å. Although both models exhibited the same overall structure, even the relative positioning of the helices was different demonstrating both the utility and the limitations of homology modeling. The total buried surface area of each molecule was roughly 1050 Å2 and spanned the length of the partial repeats. As mentioned, the N-terminal portion of α0-1 was not observed in the crystal structure. Because of its proximity to the A/B loop of β17, yet still being disordered, there is no evidence to suggest a stable interaction between the N-terminus of α-spectrin with the C-terminal β-spectrin repeats. It should be noted, that the protease-sensitive C-terminal tail of β-spectrin (residues 2085-2137) was not included because of its instability and could potentially interact with the N-terminal portion of the α chain. Nonetheless, experimental evidence to date indicates the fragments observed in the crystal structure are those primarily responsible for tetramer formation.43
Electrostatic surfaces
Although most of the contacts between the 2 molecules are hydrophobic in nature, an analysis of the electrostatic surfaces (Figure 3C) indicates that the opposing faces of the interaction do show an overall complementarity of charge. Careful examination of the calculated electrostatic map, as well as the sequence of the interacting regions, shows an enrichment of acidic residues on the surface of the β chain (Glu2022, Glu2029, Glu2045, and Glu2055). Conversely, the central portion of the α chain is rich in basic residues (Arg28, Arg34, Arg41, Arg45, and Lys48). Most of these charged residues do not form salt bridges, and their electrostatic interactions in the structure are modest compared with the predominant hydrophobic interactions. However, the overall charges of these regions may serve to guide the 2 chains into place, with the structure of the tetramerization domain being “locked” through extensive hydrophobic interactions.
Discussion
Comparison with other structures of α-spectrin repeats 0-1
Recent structural work on α-spectrin repeats 0 to 1 has provided molecular models of both the erythroid and brain isoforms by nuclear magnetic resonance (NMR)39 and crystallography, respectively (Figure 4A).42 In both the NMR39 and previous crystallographic studies,42 the isolated α0-1 domain exhibited a clear break in the helical character of the molecule. In the NMR structure (Protein Data Bank ID: 1OWA), this break occurs coincident with the linker region connecting α0 and α1; that is, a helical region corresponding to α0 is still present, but the 2 repeats seem to be folded independently.39 However, the crystal structure of the corresponding human brain α-spectrin maintains helical character for 4 additional helical turns before a secondary helical region (corresponding to the N-terminal half of brain spectrin α0). Although both structures agree well with the overall fold of repeat 1, the crystal structure of erythroid α-spectrin in the complex shows a continuous helical linker between α0 and α1. This finding suggests that although regions of α0 and its link to repeat 1 may be prone to fraying or multiple conformations in the unbound state, upon binding to the tetramerization domain of β-spectrin, these regions are stabilized. This conclusion also is supported by secondary structure prediction, which shows a marked decrease in helical propensity precisely in the sequence (GQKLED) that is disordered in the NMR structure of α0-1.39,44
In contrast to the variability in the linker region between repeats 0 and 1, the N-termini of the structures are almost uniformly disordered. Both the NMR structure of erythroid α-spectrin alone and the crystal structure of α-spectrin in complex with β-spectrin have a disordered tail of approximately 18 amino acids.39 In the NMR structure, this is demonstrated by the N-terminal tail that neither has regular secondary structure nor converges to a single solution among the multiple models deposited. In the case of our crystal structure of α-spectrin in complex with β-spectrin, the N-terminal 18 amino acids are disordered and not observed in the electron density map. In the case of brain α-spectrin, only the first 8 to 10 residues are disordered; however, in comparison with the erythroid isoform, the first 10 amino acids of brain α-spectrin are missing from the mature protein sequence. Taken together, this observation suggests that without additional interactions (which may or may not be present in vivo), the N-terminal portion of α-spectrin that precedes partial repeat 0 persists in an unstructured manner until the sequence AE(D/E)IQERR.
Comparison with other multirepeat spectrin structures
The overall structure of the α0-1/β16-17 complex displays a canonical spectrin fold consisting of triple-helical bundles connected with α-helical linkers. Although this tertiary structure had been inferred from structure prediction, biophysical measurements, and structural studies, the precise structure and positioning of α0-1 in relation to β16-17 was unknown. With the data from the current study, comparisons with both “intact” and “composite” spectrin repeat structures can now be made. Among published structures of spectrins, the majority are 1- or 2-repeat fragments.33-39,45,46 Three 3-repeat structures have been determined to date: chicken brain α-spectrin repeats 15-17, human brain β-spectrin repeats 14-16, and human erythroid β-spectrin repeats 13-15 (in complex with a fragment of ankyrin).32,45,47 Although chicken brain α-spectrin repeats 15-17 are generally linear, both human brain β-spectrin repeats 14-16 and erythroid β-spectrin repeats 13-15 display a noticeable bend of approximately 50° between repeats 14 and 15. Surprisingly, the structure of β-spectrin repeats 14-16 superimposes quite well with the 3 repeats of the complex (Figure 4B). This finding suggests that the complex formed by the tetramerization domains of α- and β-spectrin does not simply complete a 3-helix bundle—the lengths of the contributing helices, the location of the A/B loop of β-spectrin repeat 17, the presence of α-helical linkers, and the relative positioning of the 3 repeats indeed come together to form a very typical spectrin repeat.
Interestingly, the first structure of a single, spectrin-type repeat unit actually presented itself as a dimer under crystallization conditions (supplemental Figure 5).38 In many ways, the interactions seen in that structure resemble the structure of the tetramerization domain because both structures form a composite spectrin bundle from 2 molecules that maintain the length and twist found in intact spectrins.
Conservation analysis
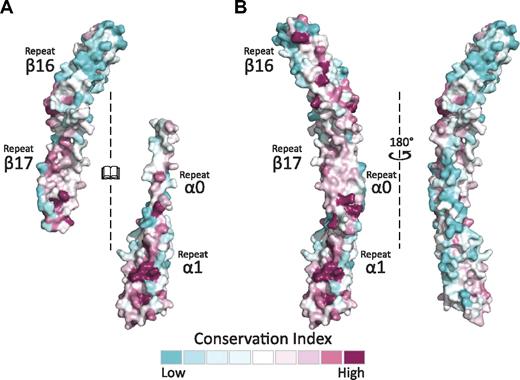
Sequence alignment and conservation mapping across species, tissue-specific isoforms, and alternative splicing variants by the use of the Consurf server28 onto the structure of both the α- and β-spectrin tetramerization domains (Figure 5A)28 indicate that, as expected, the interacting surface is more highly conserved than surfaces that do not participate in binding for these partial repeats. It should be noted that repeat β17 is among the highest conserved across β-spectrins.48 This analysis also indicates that the β-spectrin fragment maintains a greater degree of conservation than α, which supports the hypothesis that differences among α-spectrins are predominantly responsible for the varied affinities observed for different isoforms.31 Interestingly, the same face of α0 that is involved in binding β17 maintains high conservation in repeat α1 and β16 (Figure 5B). When assembled as a complex, part of the highly conserved surface is buried by the interaction. Nonetheless, an entire surface of highly conserved residues is presented on one face of the complex, suggesting that it is a biologically important, conserved surface involved in additional interactions that are currently not defined.
The spectrin tetramerization domains show multiple, highly conserved surfaces. (A) The interacting surfaces of α0-1 and β16-17 show regions of relatively high conservation. This is especially notable in comparing repeat β16 (mostly blue) to the partial repeat β17 (mostly red). As in previous figures, the interacting surfaces of both molecules are opened to face the reader. (B) Conservation analysis of the assembled tetramerization domain displays one surface with markedly greater conservation than the other, which may present a biologically relevant interaction surface. The conservation maps presented were generated using sequences and alignments derived by the Consurf database.28 An additional analysis with manually selected and aligned sequences yielded similar findings.
The spectrin tetramerization domains show multiple, highly conserved surfaces. (A) The interacting surfaces of α0-1 and β16-17 show regions of relatively high conservation. This is especially notable in comparing repeat β16 (mostly blue) to the partial repeat β17 (mostly red). As in previous figures, the interacting surfaces of both molecules are opened to face the reader. (B) Conservation analysis of the assembled tetramerization domain displays one surface with markedly greater conservation than the other, which may present a biologically relevant interaction surface. The conservation maps presented were generated using sequences and alignments derived by the Consurf database.28 An additional analysis with manually selected and aligned sequences yielded similar findings.
Insights into the mechanism of tetramerization domain binding
Comparison of the various structures of α0-1 suggests that in solution, partial repeat α0 persists as an independent helix connected to repeat α1 with a flexible linker. In contrast, the structure of α0-1 in complex with β16-17 displays a rigid, α-helical linker connecting α0 to α1. As suggested by solution studies by Long et al,31 this change in conformation likely manifests itself as a stabilization and homogenization of the varied solution states of α0-1. A more thorough analysis of the biophysical properties of the binding interaction can be extracted from both the structures discussed previously and previous determination of the thermodynamic binding parameters.17 First, on the basis of earlier measurements of the thermodynamic binding parameters (ΔH = −33.1 ± 3.5 kcal/mole; −TΔS = 24.5 ± 3.4 kcal/mole; ΔG23°C = −8.6 kcal/mole; and KD = 400 ± 100nM),17 the entropy of the complex is decreased relative to that of the individual components. This finding suggests that the increase in entropy (mostly attributable to water being excluded by the formation of the hydrophobic interface of the complex) is not sufficient to fully overcome the entropic costs of orderly forming a stable complex and limiting the flexibility of the component subunits. The assessment that such reduced molecular flexibility occurs is supported by the observation of multiple conformations in the NMR structure of α0-1.39 Nonetheless, in concert with the endothermic, salt bridge, and hydrogen bond-forming reactions, a sufficient favorable enthalpy is generated to allow formation of the complex.
Mapping of clinical mutations
The structure of the red cell spectrin α0-1/β16-17 complex enables us to begin to address the physical basis for many clinically relevant, disease-related mutations. A large number of point mutations that result in HE or HPP map to the partial repeats forming the tetramerization interface.5 In addition, mutations that truncate β-spectrin prematurely also can result in these disease states.5
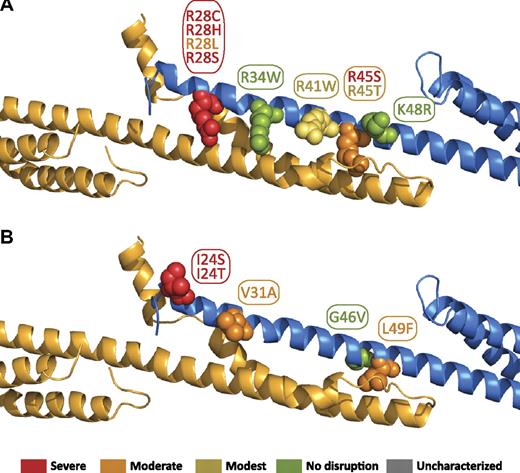
The tetramer binding affinities for known HE/HPP mutations that map to the α0 region have previously been analyzed.17 The locations of these mutations in the tetramer interface are shown in Figure 6 17 and are color-coded on the basis of the previously observed decrease in tetramer binding affinity. As has been noted previously, the majority of HE/HPP mutations in this region eliminate or modify a positively charged residue (R28[C/H/L/S], R34W, R41W, R45[S/T], and K48R). All 4 R28 mutations extensively perturb binding affinity. Consistent with this, the interaction map (supplemental Figure 4) shows this arginine participates in both hydrophobic and electrostatic interactions (with A2018 and E2022, respectively). This dual role for arginine has been observed in other spectrin structures where the aliphatic portion of an arginine side chain makes extensive hydrophobic contacts, whereas the charged, distal portion of the molecule is able to form hydrogen bonds, electrostatic interactions, or salt bridges. R41W shows a less-dramatic, but nonetheless substantial, perturbation of binding (∼ 20-fold weaker than wild type), consistent with its electrostatic and hydrophobic interactions with β17 residues.
Locations of α0 domain HE/HPP mutations in the tetramer complex. Mutations that have been associated with HE or HPP are indicated as space-filling spheres on the ribbon diagram. Most of the mutations map directly to interacting residues. The relationship to our previously reported binding affinities with the use of isothermal titration calorimetry are color coded by the use of the previously defined binding affinity categories17 : red = severe (marginal or no detected binding); orange = moderate (∼100-fold weaker than wild type); yellow = modest (∼10-fold weaker than wild type); green = no effect (wild-type binding). (A) Mutations of positively charged residues. (B) Mutations of uncharged residues.
Locations of α0 domain HE/HPP mutations in the tetramer complex. Mutations that have been associated with HE or HPP are indicated as space-filling spheres on the ribbon diagram. Most of the mutations map directly to interacting residues. The relationship to our previously reported binding affinities with the use of isothermal titration calorimetry are color coded by the use of the previously defined binding affinity categories17 : red = severe (marginal or no detected binding); orange = moderate (∼100-fold weaker than wild type); yellow = modest (∼10-fold weaker than wild type); green = no effect (wild-type binding). (A) Mutations of positively charged residues. (B) Mutations of uncharged residues.
Surprisingly, R34W shows essentially wild-type binding affinity despite electrostatic interactions with glutamic acids and hydrophobic interactions on β17. Although such a change would require minor rearrangements of the packing of the side chains in the tetramerization interface, the added hydrophobic character of the tryptophan may compensate for the loss of electrostatic interactions. In addition, hydrogen bond interactions with the indole NH may contribute to maintaining a favorable enthalpy for the binding reaction, as was measured by ITC. In addition, this side chain is partially exposed, and it is very likely that the bulkier tryptophan protrudes from the surface of the complex. The remaining arginine mutations, R45S and R45T, exhibit substantial differences in binding affinity (KD > 200 and 14μM, respectively).
On the basis of the interactions observed in the structure, it is somewhat surprising that such a moderate structural change leads to a drastic effect on binding. However, given that R45 makes extensive hydrophobic contacts, it is possible that the larger and slightly more hydrophobic threonine is able to better maintain these contacts than serine. Biophysical measurements indicate that the R45T mutation leads to a less-favorable change in enthalpy (likely because of the abolishment of the hydrogen bond interaction with D2036), whereas the entropy change is quite similar to that observed in the wild-type protein. Finally, the K48R mutation also exhibits wild-type binding, presumably because aliphatic portions of the lysine and arginine preserve normal hydrophobic interactions, whereas the charged distal portion of the side chain is exposed on the surface of the complex. The frequent loss of a positive charge in HE/HPP mutations is quite interesting because most of the contacts in the tetramer binding site are hydrophobic. This finding suggests the hypothesis that the lost charge affects long-range electrostatically driven initial recognition, thereby adversely affecting kon. The reduced kon, combined with perturbation of hydrophobic interactions that all of the arginine residues in question participate in, is likely to result in the drastic change in affinity observed for many of these mutations.
Other mutations in this portion of α-spectrin include I24(S/T), V31A, G46V, and L49F (Figure 6B). Although no direct contacts were observed in the structure involving I24, this region of the structure is close to the disordered N-terminal tail of α-spectrin, making precise placement of the I24 side chain difficult at this resolution. In addition, because this portion of the structure is in proximity to the C-terminal tail of β-spectrin, interactions of I24 with longer β-spectrin constructs may occur. The mutation of V31A, although structurally conservative, would eliminate some hydrophobic contacts and weaken the binding modestly, which is consistent with biophysical measurements indicating a more unfavorable change in entropy. Finally, both the G46V and L49F mutations likely cause local structural perturbations as the result of substitution with comparatively bulkier residues that affect the sterics of the binding interface. Consistent with the interactions evident in the structure of the complex, the authors of previous studies have proposed that these mutations affect the binding interface (G46V)49 or α-spectrin itself (L49F).50
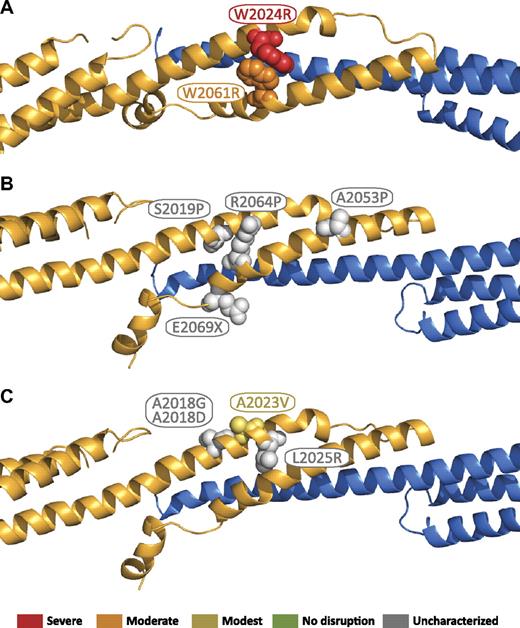
The locations of β17 mutations in the tetramer interface are shown in Figure 7.51 These mutations can be grouped into 2 general categories: those that perturb the structural integrity of the β17 domain, and those that disrupt interchain contacts. In the structure, there is considerable contact between W2024 and W2061 (Figure 7A). Changing the character of these closely packed side chains is predicted to substantially perturb the tetramer interface, and therefore, it is not surprising that W2024R, and W2061R dramatically disrupt tetramer binding, as previously reported by Nicolas et al.51 A second group of mutations that are expected to severely perturb the local structure of β17 include S2019P, A2053P, and R2064P (Figure 7B). These 3 residues do not directly interact with α-spectrin; however, their proximity to residues that interact is expected to make mutation of these residues in helical regions to proline highly detrimental. Finally, a number of premature truncations of β-spectrin occur in the β17 B helix. Most of these would disrupt a significant portion of the binding interface. The only clinical nonsense mutation in this region (the remainder of the characterized truncations are caused by frameshift mutations), E2069X, is near the end of the B helix, but it would remove a number of residues (2074, 2075, and 2078) that directly interact with the α0 domain. Finally, several of the remaining mutations are expected to disrupt specific β17-α0 interactions, including A2018G, A2018D, and L2025R (Figure 7C). In the presented structure, these residues form direct hydrophobic contacts with α-spectrin, and therefore all 3 mutations are expected to disrupt this interaction either by eliminating hydrophobic interactions, as in A2018G, or substituting a comparatively bulky, charged residue, as in A2018D and L2025R. It is less apparent that A2023V would disrupt either the β17 structure or its interaction with α0 as it is on the outside of the bundle, which is consistent with the previous findings that showed the binding affinity for this mutant was nearly that observed for the wild-type protein.51
Locations of β17 domain HE/HPP mutations in the tetramer complex. Mutations that have been associated with HE or HPP are indicated as space-filling spheres on the ribbon diagram. Where known, the relationship to reported binding affinities51 are color coded by the use of binding affinity categories similar to those used in Gaetani et al17 : red = severe (marginal or no detected binding); orange = moderate (∼ 100-fold weaker than wild type); yellow = modest (∼ 10-fold weaker than wild type); green = no effect (wild-type binding); gray = not characterized. (A) Mutations that disrupt the local structure of the β17 domain. (B) Mutations predicted to disrupt the local structure of the β17 domain. (C) Mutations predicted to disrupt association of the β17 domain with the α0 domain.
Locations of β17 domain HE/HPP mutations in the tetramer complex. Mutations that have been associated with HE or HPP are indicated as space-filling spheres on the ribbon diagram. Where known, the relationship to reported binding affinities51 are color coded by the use of binding affinity categories similar to those used in Gaetani et al17 : red = severe (marginal or no detected binding); orange = moderate (∼ 100-fold weaker than wild type); yellow = modest (∼ 10-fold weaker than wild type); green = no effect (wild-type binding); gray = not characterized. (A) Mutations that disrupt the local structure of the β17 domain. (B) Mutations predicted to disrupt the local structure of the β17 domain. (C) Mutations predicted to disrupt association of the β17 domain with the α0 domain.
In conclusion, this crystal structure of the spectrin tetramerization domain complex now provides novel insights into this critical, dynamic red cell membrane skeleton assembly site. The structure illustrates the atomic features that allow this interaction to occur and provides evidence for conformational changes in α-spectrin in response to tetramer formation. The structure of this assembly shows a striking resemblance to intact spectrin tri-repeats and also provides the first structure of the tetramerization domain of β-spectrin. A detailed analysis of the molecular surfaces affords novel insights into the nature of many HE/HPP-related mutations, whereas conservation analysis suggests additional surfaces that may form additional important interactions. This work reveals some of the key features important for spectrin tetramer formation and provides a guide for designing further experiments to explore more of the details of spectrin assembly.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We acknowledge staff and instrumentation support from the Center for Structural Biology at Northwestern University, X29 at the National Synchrotron Light Source at Brookhaven National Laboratories, and the Life Sciences Collaborative Access Team at the Advanced Photo Source at Argonne National Laboratories.
This work was supported by National Institutes of Health grants HL38794 (to D.W.S.) and GM057692 (to A.M.) and in part by an institutional grant to The Wistar Institute (CA10815).
National Institutes of Health
Authorship
Contribution: J.J.I. designed and performed research, analyzed and interpreted data, and drafted the manuscript; S.L.H. designed and performed research, analyzed and interpreted data, and assisted with manuscript preparation; T.E.M. performed research and analyzed and interpreted data; R.M. analyzed and interpreted data; and A.M. and D.W.S. designed and directed research, assisted with data interpretation, and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for T.E.M. is Vironika LLC, Philadelphia, PA.
Correspondence: Alfonso Mondragón, Department of BMBCB, Northwestern University, 2205 Tech Dr, Evanston, IL 60208; e-mail: a-mondragon@northwestern.edu; or David W. Speicher, The Wistar Institute, 3601 Spruce St, Philadelphia, PA 19104; e-mail: speicher@wistar.org.
References
Author notes
J.J.I. and S.L.H. contributed equally to this work.