Abstract
Many B-cell chronic lymphocytic leukemia (CLL) monoclonal antibodies (mAbs) can be grouped into subsets based on nearly identical stereotyped sequences. Subset 6 CLL mAbs recognize nonmuscle myosin heavy chain IIA (MYHIIA). Herein, we report that during apoptosis, MYHIIA becomes exposed on the cell surface of a subgroup of apoptotic cells, allowing subset 6 CLL mAbs to bind with it. Because other non–subset 6 CLL mAbs interact with apoptotic cells, 26 CLL mAbs, including 24 not belonging to subset 6, were tested for reactivity with MYHIIA-exposed apoptotic cells (MEACs). More than 60% of CLL mAbs bound MEACs well; most of these mAbs expressed unmutated IGHV (15 of 16) and belonged to a stereotyped subset (14 of 16). Binding to MEACs inversely correlated with the degree of IGHV mutation. Interestingly, high binding to MEACs significantly correlated with poor patient survival, suggesting that the basis of IGHV mutation status as a CLL prognostic factor reflects antigen binding. Finally, natural antibodies from human serum also reacted with MEACs. Taken together, our data indicate that a large proportion of CLL clones emerge from natural antibody-producing cells expressing immunoglobulins that recognize MEACs, and that this reactivity is associated with poor clinical outcome.
Introduction
B-cell chronic lymphocytic leukemia (CLL) is the most common Western adult leukemia, with an estimated 15 490 new cases and 4390 deaths occurring in the United States in 2009.1 CLL is a clonal expansion of CD5+CD19+ B-lymphocytes expressing a unique monoclonal antibody (mAb) that serves as the clone's B-cell antigen receptor (BCR). The amount of somatic mutation in this unique mAb predicts clinical outcome; patients with unmutated BCRs tend toward more aggressive disease.2,3 Furthermore, BCR gene sequences are virtually identical (stereotyped) in subgroups of CLL patients, with nearly 30% of patients expressing stereotyped BCRs.4 This observation suggests that a restricted set of some common antigen(s) reactive with CLL BCRs are important for the development and expansion of this disease.5,6
Previously, we identified nonmuscle myosin heavy chain IIA (MYHIIA) as an autoantigen that is recognized by subset 6 CLL mAbs.7 Subset 6 mAbs have a characteristic heavy (H) chain complementarity-determining region 3 (CDR3) sequence involving a rearrangement of unmutated IGHV1-69, IGHD3-16, and IGHJ3 that is paired with a light (L) chain with a characteristic CDR3 sequence generally involving a rearrangement of unmutated IGKV3-20.8,9 At least 53 patients with CLL worldwide share this stereotypic CLL BCR,4,7,10 suggesting that MYHIIA is a common autoreactivity among CLL clones.
MYHIIA is a large intracellular cytoplasmic protein that functions in cell shape and movement.11 In order for the CLL BCR to interact with MYHIIA, we hypothesized that MYHIIA becomes exposed on the cell surface during apoptosis.7 This idea is supported by the observations that MYHIIA is cleaved and translocates intracellularly during apoptosis,12 and that subset 6 CLL mAbs can recognize apoptotic cells.13
The studies reported here demonstrate that MYHIIA becomes exposed on the cell surface in a subgroup of apoptotic cells, and that subset 6 CLL mAbs recognize these MYHIIA-exposed apoptotic cells (MEACs). Furthermore, consistent with the findings that other CLL mAbs (not from subset 6) recognize apoptotic cells13,14 and other autoantigens7,14 that can also be exposed during apoptosis,15,16 we find that most CLL mAbs tested (16 of 26) bound MEACs. Of note, this reactivity correlates inversely with the duration of patient survival.
Finally, because binding to apoptotic cells is a characteristic of human serum natural antibodies that are generally autoreactive17,18 —properties in common with CLL mAbs7,13,14,19-23 —we now report that human serum natural antibodies bind to MEACs, supporting the idea that at least a subset of CLL clones could originate from a B-cell compartment that produces natural antibodies.
Methods
Cell culture
A human T-cell line (Jurkat) was cultured at 2 × 105 cells/mL in RPMI 1640 (Mediatech) supplemented with 10% heat inactivated fetal bovine serum (FBS; Atlanta Biologicals), 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM glutamine (Invitrogen) at 37°C and 5% CO2. Spontaneous apoptosis of Jurkat cells increased with time in culture. Starting at 2 × 105 cells/mL, 6%, 13%, 27%, 46%, and 65% Annexin V+ cells were detected after 3, 4, 5, 6, and 7 days of culture, respectively. Starting at a higher cell density increased the rate of spontaneous apoptosis (C.C.C., N.C., and L.Z., unpublished data, August-December 2008).
Patients with CLL and antibodies
Characteristics of 26 individual human patient CLL mAbs, which were prepared as recombinant human IgG1 as previously described,13,22 are listed in Table 1. Corresponding clinical information was collected from patients with CLL after informed consent as approved by the Institutional Review Board of North Shore University Hospital (Manhasset, NY) and Long Island Jewish Medical Center (New Hyde Park, NY) and in accordance with the Helsinki Declaration (Table 2). Survival times were calculated from time of first CLL diagnosis and death dates as of May 1, 2009. Kaplan-Meier survival plots were calculated using Prism (GraphPad Software).
Characteristics of CLL mAbs
| CLL no. . | Subset . | Mutation status . | % mutated . | IGHV . | HCDR3 . | IGH, GenBank accession no. . | IGKV/IGLV . | IGK/IGL, GenBank accession no. . | MFIR . |
|---|---|---|---|---|---|---|---|---|---|
| 169 | — | M | 9.0 | 3-33 | CAREGGVTGQGGFDYW | AY055480 | K4-01 | FJ039794 | 0.85 |
| 255 | — | M | 4.2 | 4-59 | CARHRGYESSGYYSSYFDYW | FJ039782 | L3-01 | FJ039804 | 1.05 |
| 141 | — | U | 0.0 | 4-34 | CARGDWRIVVVPAAVDTAMAANWFDPW | AF022005 | K1-27 | AY043160 | 1.10 |
| 260 | — | U | 0.0 | 4-b | CARAEIVVVPAAYYYYYGMDVW | FJ039783 | K1-27 | FJ039796 | 2.86 |
| 376 | — | U | 0.0 | 1-24 | CATSAFTVTHAEYFQHW | FJ039787 | K3-11 | FJ039798 | 0.89 |
| 415 | — | U | 0.0 | 1-03 | CARRPESGYSFVTPFDYW | FJ039788 | K1D-39 | FJ039799 | 1.95 |
| 562 | — | U | 1.7 | 3-21 | CVRDEITVAATRPCPW | FJ039789 | L3-21 | FJ039805 | 1.09 |
| 282 | 2 | M | 2.4 | 3-21 | CARDANGMDVW | AY553643 | L3-21 | AY574941 | 0.99 |
| 412 | 2 | U | 1.8 | 3-21 | CARDQNGMDVW | AY553648 | L3-21 | AY574946 | 1.15 |
| 183 | 4 | M | 3.2 | 4-34 | CARGYGDTPTIRRYYYYGMDVW | AF021948 | K2-30 | AY043097 | 0.74 |
| 240 | 4 | M | 3.2 | 4-34 | CARGYADTPVFRRYYYYGMDVW | AY553641 | K2-30 | AY574937 | 1.01 |
| 342 | 4 | M | 2.8 | 4-34 | CARGWGDTPMLKRYYYYGLDVW | AY553646 | K2-30 | AY574944 | 0.72 |
| 154 | 1 | M | 2.1 | 1-18 | CAREQWLVLSHFDYW | AF022009 | K1D-39 | AY043164 | 5.11 |
| 270 | 1 | U | 0.0 | 1-02 | CARVQWLGLRHFDYW | AY055487 | K1D-39 | AY574940 | 2.40 |
| 340 | 1 | U | 0.0 | 1-02 | CAREQWLVLKNFDYW | AY553645 | K1D-39 | AY574943 | 3.99 |
| 360 | 1 | U | 0.0 | 1-03 | CAREQWLVLNYFDYW | AY553647 | K1D-39 | AY574945 | 4.17 |
| 068 | 6 | U | 0.0 | 1-69 | CARGGDYDYVWGSYRSNDAFDIW | AY553640 | K3-20 | AY574935 | 3.82 |
| 258 | 6 | U | 0.0 | 1-69 | CARGGIYDYVWGSYRPNDAFDIW | AY055485 | K3-20 | AY574938 | 2.07 |
| 114 | 8 | U | 0.0 | 4-39 | CARRFGYSSSWYGLDWFDPW | AY268372 | K1D-39 | AY043094 | 17.05 |
| 657 | 8 | U | 0.3 | 4-39 | CASKTGYSSSWYGRDWFDPW | FJ039790 | K1D-39 | FJ039800 | 23.90 |
| 845 | 8 | U | 0.0 | 4-39 | CASSTGYSSSWYSPTNWFDPW | FJ039791 | K1D-39 | FJ039801 | 25.25 |
| 014 | 9 | U | 0.0 | 1-69 | CATKNDFWSGYYEGYYYYYYMDVW | AF021951 | L3-01 | AY043106 | 1.84 |
| 246 | 9 | U | 0.3 | 1-69 | CARSDQNYDFWSGYFRYYGMDVW | FJ039781 | K3-20 | FJ039795 | 9.53 |
| 355 | 9 | U | 0.0 | 1-69 | CARADLPYYDFWSGMYYYGMDVW | FJ039784 | K1-05 | AJ697904 | 22.55 |
| 366 | 9 | U | 0.0 | 3-21 | CARGVLNYDFWSVYYYYGMDVW | FJ039786 | K4-01 | FJ039797 | 3.56 |
| DO13 | 28 | U | 0.0 | 1-02 | CARQFSGSPTRYYYYYGMDVW | FJ039793 | K4-01 | FJ039803 | 10.08 |
| CLL no. . | Subset . | Mutation status . | % mutated . | IGHV . | HCDR3 . | IGH, GenBank accession no. . | IGKV/IGLV . | IGK/IGL, GenBank accession no. . | MFIR . |
|---|---|---|---|---|---|---|---|---|---|
| 169 | — | M | 9.0 | 3-33 | CAREGGVTGQGGFDYW | AY055480 | K4-01 | FJ039794 | 0.85 |
| 255 | — | M | 4.2 | 4-59 | CARHRGYESSGYYSSYFDYW | FJ039782 | L3-01 | FJ039804 | 1.05 |
| 141 | — | U | 0.0 | 4-34 | CARGDWRIVVVPAAVDTAMAANWFDPW | AF022005 | K1-27 | AY043160 | 1.10 |
| 260 | — | U | 0.0 | 4-b | CARAEIVVVPAAYYYYYGMDVW | FJ039783 | K1-27 | FJ039796 | 2.86 |
| 376 | — | U | 0.0 | 1-24 | CATSAFTVTHAEYFQHW | FJ039787 | K3-11 | FJ039798 | 0.89 |
| 415 | — | U | 0.0 | 1-03 | CARRPESGYSFVTPFDYW | FJ039788 | K1D-39 | FJ039799 | 1.95 |
| 562 | — | U | 1.7 | 3-21 | CVRDEITVAATRPCPW | FJ039789 | L3-21 | FJ039805 | 1.09 |
| 282 | 2 | M | 2.4 | 3-21 | CARDANGMDVW | AY553643 | L3-21 | AY574941 | 0.99 |
| 412 | 2 | U | 1.8 | 3-21 | CARDQNGMDVW | AY553648 | L3-21 | AY574946 | 1.15 |
| 183 | 4 | M | 3.2 | 4-34 | CARGYGDTPTIRRYYYYGMDVW | AF021948 | K2-30 | AY043097 | 0.74 |
| 240 | 4 | M | 3.2 | 4-34 | CARGYADTPVFRRYYYYGMDVW | AY553641 | K2-30 | AY574937 | 1.01 |
| 342 | 4 | M | 2.8 | 4-34 | CARGWGDTPMLKRYYYYGLDVW | AY553646 | K2-30 | AY574944 | 0.72 |
| 154 | 1 | M | 2.1 | 1-18 | CAREQWLVLSHFDYW | AF022009 | K1D-39 | AY043164 | 5.11 |
| 270 | 1 | U | 0.0 | 1-02 | CARVQWLGLRHFDYW | AY055487 | K1D-39 | AY574940 | 2.40 |
| 340 | 1 | U | 0.0 | 1-02 | CAREQWLVLKNFDYW | AY553645 | K1D-39 | AY574943 | 3.99 |
| 360 | 1 | U | 0.0 | 1-03 | CAREQWLVLNYFDYW | AY553647 | K1D-39 | AY574945 | 4.17 |
| 068 | 6 | U | 0.0 | 1-69 | CARGGDYDYVWGSYRSNDAFDIW | AY553640 | K3-20 | AY574935 | 3.82 |
| 258 | 6 | U | 0.0 | 1-69 | CARGGIYDYVWGSYRPNDAFDIW | AY055485 | K3-20 | AY574938 | 2.07 |
| 114 | 8 | U | 0.0 | 4-39 | CARRFGYSSSWYGLDWFDPW | AY268372 | K1D-39 | AY043094 | 17.05 |
| 657 | 8 | U | 0.3 | 4-39 | CASKTGYSSSWYGRDWFDPW | FJ039790 | K1D-39 | FJ039800 | 23.90 |
| 845 | 8 | U | 0.0 | 4-39 | CASSTGYSSSWYSPTNWFDPW | FJ039791 | K1D-39 | FJ039801 | 25.25 |
| 014 | 9 | U | 0.0 | 1-69 | CATKNDFWSGYYEGYYYYYYMDVW | AF021951 | L3-01 | AY043106 | 1.84 |
| 246 | 9 | U | 0.3 | 1-69 | CARSDQNYDFWSGYFRYYGMDVW | FJ039781 | K3-20 | FJ039795 | 9.53 |
| 355 | 9 | U | 0.0 | 1-69 | CARADLPYYDFWSGMYYYGMDVW | FJ039784 | K1-05 | AJ697904 | 22.55 |
| 366 | 9 | U | 0.0 | 3-21 | CARGVLNYDFWSVYYYYGMDVW | FJ039786 | K4-01 | FJ039797 | 3.56 |
| DO13 | 28 | U | 0.0 | 1-02 | CARQFSGSPTRYYYYYGMDVW | FJ039793 | K4-01 | FJ039803 | 10.08 |
Subset number or — indicates not attributable to a defined subset.4,13,24 U indicates unmutated; and M, more than 2% mutated compared with germline according to IMGT.25
IGHV mutation percentage compared with germline according to IMGT.25 IGHV, IGKV (K), and IGLV (L) gene name is according to IMGT nomenclature.25
HCDR3 amino acid sequence is shown in single-letter code plus junctional C and W residues according to IMGT.25
MFIR measurement of MEAC binding from Figure 3C.
Characteristics of patients with CLL
| CLL no. . | Sex . | ZAP-70, % positive . | CD38, % positive . | Age at diagnosis, y . | Survival after diagnosis, mo* . |
|---|---|---|---|---|---|
| 169 | M | 0.5 | 5.1 | 34 | > 153 |
| 255 | M | 20.9 | 2.4 | 57 | > 202 |
| 141 | M | 35.5 | 88.4 | 76 | 99 |
| 260 | F | 16.0 | 33.2 | 69 | 163 |
| 376 | F | ND | ND | 76 | > 166 |
| 415 | M | ND | 99.4 | 60 | > 85 |
| 562 | M | 32.1 | 70.6 | 49 | 75 |
| 282 | M | ND | ND | 61 | > 115 |
| 412 | M | ND | 15.8 | 53 | > 175 |
| 183 | M | 15.7 | 7.3 | 63 | > 145 |
| 240 | F | 13.4 | 69.7 | — | — |
| 342 | F | 1.0 | 2.2 | 56 | > 234 |
| 154 | M | 15.5 | 24.9 | 66 | 59 |
| 270 | M | 94.0 | 76.4 | 55 | > 163 |
| 340 | F | ND | ND | 41 | 95 |
| 360 | M | ND | 31.4 | 57 | > 117 |
| 068 | M | 52.6 | 45.9 | 65 | 151 |
| 258 | M | 40.7 | 3.0 | 65 | 99 |
| 114 | M | ND | ND | 59 | 53 |
| 657 | M | 26.3 | 84.5 | 62 | 53 |
| 845 | M | 42.5 | 96.7 | 75 | > 41 |
| 014 | M | 98.1 | 98.0 | 59 | 145 |
| 246 | M | 11.5 | 94.2 | 75 | 20 |
| 355 | M | 11.7 | 22.4 | 65 | 118 |
| 366 | M | ND | ND | 73 | 42 |
| DO13 | F | ND | ND | — | — |
| CLL no. . | Sex . | ZAP-70, % positive . | CD38, % positive . | Age at diagnosis, y . | Survival after diagnosis, mo* . |
|---|---|---|---|---|---|
| 169 | M | 0.5 | 5.1 | 34 | > 153 |
| 255 | M | 20.9 | 2.4 | 57 | > 202 |
| 141 | M | 35.5 | 88.4 | 76 | 99 |
| 260 | F | 16.0 | 33.2 | 69 | 163 |
| 376 | F | ND | ND | 76 | > 166 |
| 415 | M | ND | 99.4 | 60 | > 85 |
| 562 | M | 32.1 | 70.6 | 49 | 75 |
| 282 | M | ND | ND | 61 | > 115 |
| 412 | M | ND | 15.8 | 53 | > 175 |
| 183 | M | 15.7 | 7.3 | 63 | > 145 |
| 240 | F | 13.4 | 69.7 | — | — |
| 342 | F | 1.0 | 2.2 | 56 | > 234 |
| 154 | M | 15.5 | 24.9 | 66 | 59 |
| 270 | M | 94.0 | 76.4 | 55 | > 163 |
| 340 | F | ND | ND | 41 | 95 |
| 360 | M | ND | 31.4 | 57 | > 117 |
| 068 | M | 52.6 | 45.9 | 65 | 151 |
| 258 | M | 40.7 | 3.0 | 65 | 99 |
| 114 | M | ND | ND | 59 | 53 |
| 657 | M | 26.3 | 84.5 | 62 | 53 |
| 845 | M | 42.5 | 96.7 | 75 | > 41 |
| 014 | M | 98.1 | 98.0 | 59 | 145 |
| 246 | M | 11.5 | 94.2 | 75 | 20 |
| 355 | M | 11.7 | 22.4 | 65 | 118 |
| 366 | M | ND | ND | 73 | 42 |
| DO13 | F | ND | ND | — | — |
M indicates male; F, female; ND, not determined; and —, not known.
> indicates patient is still alive as of May 1, 2009.
Fluorescence microscopy
Jurkat cell cultures containing T cells undergoing spontaneous apoptosis were resuspended at 2 × 104 cells/mL in phosphate-buffered saline (PBS; Mediatech) and 500 μL of cell suspension were placed on poly-L-lysine–coated glass coverslips (BD Biosciences) in 24-well flat-bottom tissue-culture plates (BD Biosciences). Cells were fixed with 4% formaldehyde (Ted Pella Inc) in PBS for 10 minutes at room temperature (RT) and washed twice for 5 minutes in PBS. To block nonspecific binding, coverslips were incubated in 5% FBS in PBS for 30 minutes at RT in a dark humidifying chamber. Coverslips were incubated with 35 μL of CLL mAb (150-200 μg/mL) and/or 35 μL of rabbit anti–human MYHIIA antibody (1:50, BTI-561; Biomedical Technologies) diluted in 5% FBS in PBS for 1 hour at RT, washed 3 times in PBS for 5 minutes, incubated with 1 drop fluorescein isothiocyanate (FITC)–conjugated anti–human IgG (INOVA Diagnostics) and/or 35 μL (6 μg/mL) of Rhodamine Red-X or (7.5 μg/mL) FITC-conjugated donkey anti–rabbit IgG (Jackson ImmunoResearch Laboratories) diluted in 5% milk in PBS for 30 minutes at RT, and washed 3 times in PBS for 5 minutes. For analysis of DNA condensation during apoptosis, coverslips were further incubated with 0.25 μg/mL propidium iodide (PI; BD Biosciences) in PBS for 10 minutes at RT and washed 3 times in PBS for 5 minutes. Coverslips were mounted on Colorfrost Plus microscope slides (Thermo Fisher Scientific) with 70% VECTASHIELD Mounting Medium (Vector Laboratories) and sealed with nail polish for 15 minutes. Slides were imaged by a confocal laser-scanning microscope system (FluoView 300-IX; Olympus) using a PLAN APO 60×/1.4 oil-objective lens. Representative apoptotic cells were chosen based on nuclear fragmentation detected by PI staining or based on subsequent MYHIIA+ staining consistent with rearrangement during apoptosis. Images were processed and analyzed using Adobe Photoshop CS3 (Adobe Systems Inc) and ImageJ with an intensity correlation analysis plug-in (National Institutes of Health; http://www.macbiophotonics.ca/imagej/) to statistically evaluate colocalization.26,27 Intensity correlation analysis not only measures the color overlap between 2 overlain images, but also tests if the intensities of the 2 colors vary in synchrony, as one would expect if the colors tagged the same molecule.27 Default thresholds were used and the resulting Pearson correlation coefficient (r), which varies from −1 to 1, where 1 equals complete colocalization, is reported.
Flow cytometry
Cells were resuspended at 5 × 106 cells/mL in flow cytometry wash buffer (FWB; 1% bovine serum albumin, 0.1% sodium azide [Sigma-Aldrich] in PBS). A total of 100 μL of cell suspension was incubated with 25 to 200 μg/mL human CLL mAb, normal human serum IgG (Sigma-Aldrich or Miltenyi Biotec), humanized anti–human CD20 (human IgG1 [rituximab]; Genentech), and/or rabbit anti–human MYHIIA (BTI-564; Biomedical Technologies) for 1 hour at RT, washed 2 times in PBS, resuspended in 100 μL of FWB, and incubated with 0.2 to 2 μg/mL FITC-conjugated F(ab′)2 goat anti–human IgG (Southern Biotech) or 1 to 2.4 μg/mL phycoerythrin (PE)–conjugated goat anti–rabbit IgG (Southern Biotech), or 1 to 6 μg/mL FITC-conjugated donkey anti–rabbit IgG (Jackson ImmunoResearch Laboratories) for 30 minutes at RT. For apoptosis analysis, cells were further washed twice in PBS and once in annexin V binding buffer (AVBB; BD Biosciences), incubated with 4 μL of PE-conjugated annexin V (AV-PE; BD Biosciences) and 7-amino-actinomycin (7AAD; BD Biosciences) in 100 μL of AVBB for 15 minutes at RT, and then 400 μL of AVBB was added. Flow cytometry data were collected on the cell samples using a FACSCalibur or LSRII (BD Biosciences) and analyzed with FlowJo (TreeStar). CLL cells were phenotyped by flow cytometry as previously described.28 Briefly, CD38 and ZAP-70 percentages of positivity in CD5+ CD19+ CLL cells were determined using anti-CD38–PE (BD Biosciences) and anti–ZAP-70–FITC (eBioscience).
Results
CLL subset 6 mAb recognizes MYHIIA exposed during apoptosis
MYHIIA rearranges intracellularly during apoptosis.12 Therefore, using confocal microscopy, we examined Jurkat T cells spontaneously undergoing apoptosis for MYHIIA expression at the cell surface. This required the use of nonpermeabilizing conditions to avoid intracellular binding of anti-MYHIIA to its target. Under these conditions, anti-MYHIIA generally only bound apoptotic cells marked by nuclear DNA staining by PI (representative examples shown in Figure 1A). Apoptotic cells with evident nuclear fragmentation generally exhibited large punctate bodies containing MYHIIA that interestingly did not colocalize with condensed DNA as evidenced by the lack of overlap between anti-MYHIIA and PI staining (r = .156). Furthermore, CLL subset 6 mAb 068 exhibited the same staining pattern as anti-MYHIIA (representative examples shown in Figure 1B), confirming that CLL 068 mAb recognizes apoptotic cells.13 Like anti-MYHIIA antibodies, CLL 068 mAb binding did not colocalize with DNA condensation during apoptosis (r = .048). However, colocalization of anti-MYHIIA and CLL 068 staining was observed on apoptotic cells (representative examples shown in Figure 1C), which had large punctate bodies visualized by anti-MYHIIA and CLL 068 with appreciable overlap (r = .647). Thus, the CLL 068 subset 6 mAb recognized MYHIIA exposed during apoptosis.
Apoptosis exposes MYHIIA and permits CLL subset 6 mAb reactivity. (A-B) Spontaneous apoptosis in Jurkat cells was revealed by propidium iodide (PI; red)–stained DNA in condensed nuclei. Apoptotic cells were costained under nonpermeabilizing conditions with (A) anti-MYHIIA antibody (green) or (B) CLL 068 mAb (green) and visualized by confocal microscopy (original magnification, ×600). Two representative cells from 6 independent experiments are shown with the average Pearson correlation coefficient (r) as a measure of colocalization. (C) Spontaneous apoptotic Jurkat cells were stained under nonpermeabilizing conditions with anti-MYHIIA (red) and CLL 068 mAb (green) and visualized by confocal microscopy (original magnification, ×600). Two representative cells from 3 independent experiments are shown with the average r. In all experiments (A-C), merges of red and green panels are shown (Merge; overlap in yellow-orange) and staining with secondary antibody alone was negative (not shown).
Apoptosis exposes MYHIIA and permits CLL subset 6 mAb reactivity. (A-B) Spontaneous apoptosis in Jurkat cells was revealed by propidium iodide (PI; red)–stained DNA in condensed nuclei. Apoptotic cells were costained under nonpermeabilizing conditions with (A) anti-MYHIIA antibody (green) or (B) CLL 068 mAb (green) and visualized by confocal microscopy (original magnification, ×600). Two representative cells from 6 independent experiments are shown with the average Pearson correlation coefficient (r) as a measure of colocalization. (C) Spontaneous apoptotic Jurkat cells were stained under nonpermeabilizing conditions with anti-MYHIIA (red) and CLL 068 mAb (green) and visualized by confocal microscopy (original magnification, ×600). Two representative cells from 3 independent experiments are shown with the average r. In all experiments (A-C), merges of red and green panels are shown (Merge; overlap in yellow-orange) and staining with secondary antibody alone was negative (not shown).
CLL subset 6 mAb recognizes MYHIIA exposed on only a subset of apoptotic cells
To quantify these fluorescence microscopy observations, apoptotic cells were examined by flow cytometry to measure the levels of exposure of MYHIIA in apoptotic cells. In addition, we determined MYHIIA exposure during early and/or late apoptosis by staining with AV-PE and 7AAD, which separates live cells (AV-PE−, 7AAD−) from early (AV-PE+, 7AAD−) and late (AV-PE+, 7AAD+) apoptotic cells. Costaining Jurkat cells with AV-PE, 7AAD, and anti-MYHIIA allowed us to gate on early or late apoptotic cells and then examine the level of MYHIIA expression, revealing that only a fraction of the apoptotic cells expose MYHIIA (Figure 2A right panels). In contrast, gating on live cells revealed no MYHIIA exposure (Figure 2A bottom left panel). The lack of live cell binding is also clearly seen by gating on MYHIIA+ cells (Figure 2B top panel) and examining the AV-PE and 7AAD patterns (Figure 2B bottom panel). In this case, only the apoptotic fractions (early and late) and not the live cell fraction are identified.
MYHIIA and CLL subset 6 mAb reactivity is exposed on a subset of early and late apoptotic cells. Flow cytometric analyses of spontaneous apoptotic Jurkat cells are displayed as contour plots of fluorescence intensity shown on 4-log scales with BiExponential transformation. (A) Cells were stained with rabbit anti–human MYHIIA antibody (100 μg/mL), FITC-conjugated anti–rabbit IgG (3 μg/mL), 7AAD, and AV-PE. Representatives of 9 experiments are shown. Early apoptotic cells (7AAD−, AV-PE+, 26.3%; top left panel) were gated and contain a subset of MYHIIA+ cells (22.9%; bottom right panel). Late apoptotic cells (7AAD+, AV-PE+, 14.5%; top left panel) were gated and contain a subset of MYHIIA+ cells (9.4%; top right panel). Gated live cells (7AAD−, AV-PE−, 58.5%; top left panel) contained no MYHIIA+ cells (0.4%; bottom left panel). (B) Same experiment as in panel A showing MYHIIA+ cells (7.3%) are all AV-PE+ (top panel). Gated MYHIIA+ cells are either early (78.9%) or late (19.9%) apoptotic cells (bottom panel). (C) Cells were stained with human CLL 068 mAb (100 μg/mL), FITC-conjugated anti–human IgG (2 μg/mL), 7AAD, and AV-PE. Representatives of 23 experiments are shown. Early apoptotic cells (23.7%; top left panel) were gated and contain a subset of CLL 068+ cells (27.3%; bottom right panel). Late apoptotic cells (23.0%; top left panel) were gated and contain a subset of CLL 068+ cells (8.3%; top right panel). Gated live cells (52.4%; top left panel) contained no CLL 068+ cells (0.1%; bottom left panel). (D) Same experiment as in panel C showing CLL 068+ cells (8.7%) are all AV-PE+ (top panel). Gated CLL 068+ cells are either early (73.6%) or late (24.8%) apoptotic cells (bottom panel). (E) Cells were stained with CLL 068 mAb (50 μg/mL) plus FITC-conjugated anti–human IgG (0.2 μg/mL; x-axis) and PE-conjugated anti–rabbit IgG alone (2.4 μg/mL), AV-PE, or anti-MYHIIA antibody (200 μg/mL) plus PE-conjugated anti–rabbit IgG (y-axis; left to right, respectively). Representatives of 19 experiments are shown. In this experiment, CLL mAb 068 reacts with 36.6% cells with less than 1% background PE staining (left panel). CLL 068 only stains AV-PE+ cells (44.1%), with a subset of AV-PE+ cells negative for CLL 068 staining (19.8%; middle panel). All CLL 068+ cells are MYHIIA+ (42.8%; right panel). Cells were not stained by secondary antibodies alone (not shown).
MYHIIA and CLL subset 6 mAb reactivity is exposed on a subset of early and late apoptotic cells. Flow cytometric analyses of spontaneous apoptotic Jurkat cells are displayed as contour plots of fluorescence intensity shown on 4-log scales with BiExponential transformation. (A) Cells were stained with rabbit anti–human MYHIIA antibody (100 μg/mL), FITC-conjugated anti–rabbit IgG (3 μg/mL), 7AAD, and AV-PE. Representatives of 9 experiments are shown. Early apoptotic cells (7AAD−, AV-PE+, 26.3%; top left panel) were gated and contain a subset of MYHIIA+ cells (22.9%; bottom right panel). Late apoptotic cells (7AAD+, AV-PE+, 14.5%; top left panel) were gated and contain a subset of MYHIIA+ cells (9.4%; top right panel). Gated live cells (7AAD−, AV-PE−, 58.5%; top left panel) contained no MYHIIA+ cells (0.4%; bottom left panel). (B) Same experiment as in panel A showing MYHIIA+ cells (7.3%) are all AV-PE+ (top panel). Gated MYHIIA+ cells are either early (78.9%) or late (19.9%) apoptotic cells (bottom panel). (C) Cells were stained with human CLL 068 mAb (100 μg/mL), FITC-conjugated anti–human IgG (2 μg/mL), 7AAD, and AV-PE. Representatives of 23 experiments are shown. Early apoptotic cells (23.7%; top left panel) were gated and contain a subset of CLL 068+ cells (27.3%; bottom right panel). Late apoptotic cells (23.0%; top left panel) were gated and contain a subset of CLL 068+ cells (8.3%; top right panel). Gated live cells (52.4%; top left panel) contained no CLL 068+ cells (0.1%; bottom left panel). (D) Same experiment as in panel C showing CLL 068+ cells (8.7%) are all AV-PE+ (top panel). Gated CLL 068+ cells are either early (73.6%) or late (24.8%) apoptotic cells (bottom panel). (E) Cells were stained with CLL 068 mAb (50 μg/mL) plus FITC-conjugated anti–human IgG (0.2 μg/mL; x-axis) and PE-conjugated anti–rabbit IgG alone (2.4 μg/mL), AV-PE, or anti-MYHIIA antibody (200 μg/mL) plus PE-conjugated anti–rabbit IgG (y-axis; left to right, respectively). Representatives of 19 experiments are shown. In this experiment, CLL mAb 068 reacts with 36.6% cells with less than 1% background PE staining (left panel). CLL 068 only stains AV-PE+ cells (44.1%), with a subset of AV-PE+ cells negative for CLL 068 staining (19.8%; middle panel). All CLL 068+ cells are MYHIIA+ (42.8%; right panel). Cells were not stained by secondary antibodies alone (not shown).
In a similar fashion, subset 6 mAb 068 also recognized a subgroup of both early and late apoptotic cells, but not live cells (Figure 2C-D). Of note, the observation that live cells and a subgroup of apoptotic cells were not MYHIIA+ or CLL 068+ served as an internal control indicating that the anti-MYHIIA and CLL antibodies were not binding to cells nonspecifically. Finally, both CLL 068 mAb and anti-MYHIIA bound the same population of cells (Figure 2E right panel). CLL 068 staining alone had very little background fluorescence in the PE channel (Figure 2E left panel) and CLL 068 stained only apoptotic (AV-PE+) cells (Figure 2E middle panel). Thus, CLL 068 subset 6 mAb recognizes MYHIIA+ apoptotic cells.
Multiple CLL mAbs react with MEACs
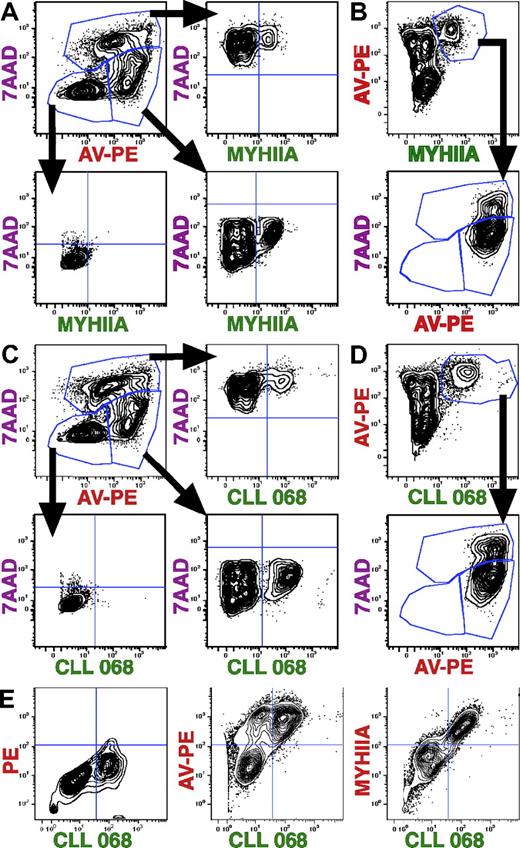
Because MYHIIA was exposed only on a subgroup of apoptotic cells, we named this fraction “MYHIIA-exposed apoptotic cells” (MEACs). As anticipated, CLL subset 6 mAb 068 recognized MEACs due to its recognition of MYHIIA.7 Because other non–subset 6 CLL mAbs are able to recognize apoptotic cells,13-16 perhaps through recognition of exposed antigens other than MYHIIA, we tested additional CLL mAbs for reactivity with MEACs (Figure 3; Table 1). As before, MYHIIA surface membrane expression was used as a marker of apoptotic cells belonging to the MEAC subgroup (Figure 2). A total of 2 CLL subset 6 mAbs (CLL 068 and CLL 258) bound MEACs (Figure 3A top right and bottom left panels) compared with apoptotic cells stained with secondary antibodies alone (Figure 3A top left panel) or with anti-MYHIIA alone (Figure 3A top middle panel). Of note, CLL mAbs not belonging to subset 6 also did (CLL 114) or did not (CLL 169) costain MEACs (Figure 3A bottom middle and right panels).
Multiple CLL mAbs react with MEACs. Spontaneous apoptotic Jurkat cells were stained with rabbit anti–human MYHIIA antibody (25 μg/mL) and CLL mAb (25 μg/mL), followed by secondary antibodies: PE-conjugated anti–rabbit IgG (1 μg/mL) and FITC-conjugated anti–human IgG (1 μg/mL). (A) Flow cytometric analyses are displayed as contour plots of fluorescence intensity shown on log scales with BiExponential transformation. Top left panel shows cells stained with secondary antibodies alone. Top middle panel shows cells stained with all reagents except CLL mAb. The remaining panels show staining with different CLL mAbs as indicated. (B) After gating on MYHIIA+ cells, histograms of the percentage of maximum (% of Max) fluorescent intensity for indicated CLL mAb staining (blue thick line) is shown relative to cells stained with all reagents except CLL mAb (red thin line). CLL 258 histogram (not shown) was similar to that of CLL 068. (C) Chart showing the MFIR staining of MEACs with 26 CLL mAbs. The MFI for CLL mAb staining of MYHIIA+ cells was determined from histograms as shown in panel B. The MFIR was calculated by dividing with the MFI obtained for MYHIIA+ cells that were stained with all reagents except CLL mAb. The CLL mAb is indicated on the x-axis. The CLL mAb subset is indicated underneath the CLL mAb numbers (subset number or “-” if not part of a subset). The bottom row of letters indicates if the IGHV of a CLL mAb is mutated (M) or not (U).
Multiple CLL mAbs react with MEACs. Spontaneous apoptotic Jurkat cells were stained with rabbit anti–human MYHIIA antibody (25 μg/mL) and CLL mAb (25 μg/mL), followed by secondary antibodies: PE-conjugated anti–rabbit IgG (1 μg/mL) and FITC-conjugated anti–human IgG (1 μg/mL). (A) Flow cytometric analyses are displayed as contour plots of fluorescence intensity shown on log scales with BiExponential transformation. Top left panel shows cells stained with secondary antibodies alone. Top middle panel shows cells stained with all reagents except CLL mAb. The remaining panels show staining with different CLL mAbs as indicated. (B) After gating on MYHIIA+ cells, histograms of the percentage of maximum (% of Max) fluorescent intensity for indicated CLL mAb staining (blue thick line) is shown relative to cells stained with all reagents except CLL mAb (red thin line). CLL 258 histogram (not shown) was similar to that of CLL 068. (C) Chart showing the MFIR staining of MEACs with 26 CLL mAbs. The MFI for CLL mAb staining of MYHIIA+ cells was determined from histograms as shown in panel B. The MFIR was calculated by dividing with the MFI obtained for MYHIIA+ cells that were stained with all reagents except CLL mAb. The CLL mAb is indicated on the x-axis. The CLL mAb subset is indicated underneath the CLL mAb numbers (subset number or “-” if not part of a subset). The bottom row of letters indicates if the IGHV of a CLL mAb is mutated (M) or not (U).
To clearly distinguish the binding to MEACs from binding to live cells, the geometric mean fluorescence intensity (MFI) staining of CLL mAbs was determined from histograms after gating on only MYHIIA+ cells (Figure 3B). CLL subset 6 mAbs (represented by CLL 068) have a higher MFI than that for cells stained with anti-MYHIIA alone (Figure 3B left panel). Other CLL mAbs binding to MEACs (CLL 114) or not (CLL 169) were discriminated in this way (Figure 3B middle and right panels).
To quantify MEAC binding for a large panel of CLL mAbs (n = 26), the MFI ratio (MFIR) of CLL mAb staining compared with anti-MYHIIA staining alone was determined (Figure 3C; Table 1). Most CLL mAbs (16 of 26) bound MEACs well (MFIR ≥ 1.5). An MFIR less than 1.5 was chosen as the cutoff for low binding because it corresponds to binding that is approximately the same as the negative control. CLL mAbs belonging to the same stereotyped subset bound MEACs in the same manner, either well (subsets 1, 6, 8, 9, and 28) or not (subsets 2 and 4). Of the stereotyped subsets, subset 8 CLL mAbs had the highest levels of binding to MEACs. Subset 9 had the most variability in MEAC binding of all the subsets; this variability may reflect the relatively diverse HCDR3 sequences of these mAbs (Table 1).
Relationship between IGHV mutation status and MEAC binding
Overall, unmutated CLL mAbs more often exhibited higher MEAC binding (15 of 19 unmutated vs 1 of 7 mutated). In addition, the one mutated CLL mAb that bound MEACs well (CLL 154) reacted similarly to other members of subset 1; note that CLL 154 only minimally exceeds the mutated IGHV cutoff (2.1%; Table 1). Of the 4 unmutated CLL mAbs that did not bind MEACs well, 2 (CLL 412 and CLL 562) also exhibited borderline unmutated IGHV status (1.8% and 1.7%, respectively; Table 1). Collectively, these findings suggest that the degree of IGHV mutation correlates with MEAC binding.
Using the exact Mann-Whitney test to examine the association of IGHV mutation status and MFIR, we found a significant association between mutation status and MFIR (P < .002). Mutated CLL mAbs had lower MFIR levels (median MFIR, 1.0; interquartile range, 0.3) than did unmutated CLL mAbs (median MFIR, 3.6; interquartile range, 8.2). Furthermore, to measure the degree of correlation of amount of MEAC binding and percentage of IGHV mutation, we computed the Spearman correlation coefficient (Figure 4A). The percentage of IGHV mutation inversely correlated with the intensity of MEAC binding in a statistically significant manner.
CLL mAb reactivity with MEACs correlates with IGHV mutation and clinical survival. (A) MEAC reactivity, measured as MFIR in Figure 3C, was plotted versus percentage of CLL mAb IGHV mutation, showing a moderate inverse correlation, where a decrease in MFIR correlated with an increase in percentage of mutation (Spearman nonparametric correlation coefficient equals −.556 with P value as shown). (B) Kaplan-Meier survival plot comparing patients with CLL with mAbs having low (Lo) versus high (Hi; MFIR ≥ 1.5) binding to MEACs. Clinical information was available for patients from 24 of 26 tested CLL mAbs. Patients with CLL whose mAb had Hi binding to MEACs (n = 15) had a median survival time of 99 months, whereas median survival for those with Lo binding (n = 9) was not reached. The statistical difference (P value) between these curves is shown. (C) Kaplan-Meier survival plot comparing the same patients on the basis of unmutated (UM) versus mutated (Mut; > 2.0%) IGHV. Patients with CLL with UM mAb (n = 18) had a median survival time of 118 months, whereas median survival for those with Mut mAb (n = 6) was not reached. The statistical difference (P value) between these curves is shown.
CLL mAb reactivity with MEACs correlates with IGHV mutation and clinical survival. (A) MEAC reactivity, measured as MFIR in Figure 3C, was plotted versus percentage of CLL mAb IGHV mutation, showing a moderate inverse correlation, where a decrease in MFIR correlated with an increase in percentage of mutation (Spearman nonparametric correlation coefficient equals −.556 with P value as shown). (B) Kaplan-Meier survival plot comparing patients with CLL with mAbs having low (Lo) versus high (Hi; MFIR ≥ 1.5) binding to MEACs. Clinical information was available for patients from 24 of 26 tested CLL mAbs. Patients with CLL whose mAb had Hi binding to MEACs (n = 15) had a median survival time of 99 months, whereas median survival for those with Lo binding (n = 9) was not reached. The statistical difference (P value) between these curves is shown. (C) Kaplan-Meier survival plot comparing the same patients on the basis of unmutated (UM) versus mutated (Mut; > 2.0%) IGHV. Patients with CLL with UM mAb (n = 18) had a median survival time of 118 months, whereas median survival for those with Mut mAb (n = 6) was not reached. The statistical difference (P value) between these curves is shown.
MEAC binding correlates with poor patient survival
Because the level of IGHV mutation correlates with aggressive disease,2,3 we analyzed the relationship between patient outcome and mAb binding to MEACs (Figure 4B; Tables 1–2). Patients whose CLL mAbs bound MEACs well (MFIR ≥ 1.5) tended to have a shorter survival than those whose mAbs had low MEAC binding. This difference was statistically significant (P < .009). Interestingly, in this patient cohort, the correlation of patient survival with CLL mAb binding to MEACs was better than that observed with IGHV mutation status (Figure 4C; Tables 1–2), which did not reach statistical significance (P < .058). MEAC binding correlated better than IGHV mutation status with survival because 1 patient with CLL (CLL 154) with a mutated IGHV CLL mAb that bound MEACs well (Figure 3C; Tables 1–2) had poor survival and 2 of 4 patients (CLL 376 and CLL 412, and not CLL 141 or CLL 562) with unmutated IGHV CLL mAb that bound MEACs poorly (Figure 3C; Tables 1–2) had longer survival times.
Using Cox proportional hazards models, we examined the effects of the degree of IGHV mutation (percentage) or MEAC binding (MFIR) on overall survival. In this analysis, only MFIR was significantly associated with survival. Patients with lower MFIR were more likely to survive (P < .011). For each unit decrease in MFIR, the patient was 1.088 times more likely to survive (95% confidence interval [CI] = 1.02-1.16). The levels of prognostic markers CD382 and ZAP-7029 in CLL cells could be measured in 17 and 20 patients, respectively (Table 2). In these cases, CD38 and ZAP-70 levels did not significantly correlate with MEAC binding or patient survival.
Natural antibodies bind MEACs
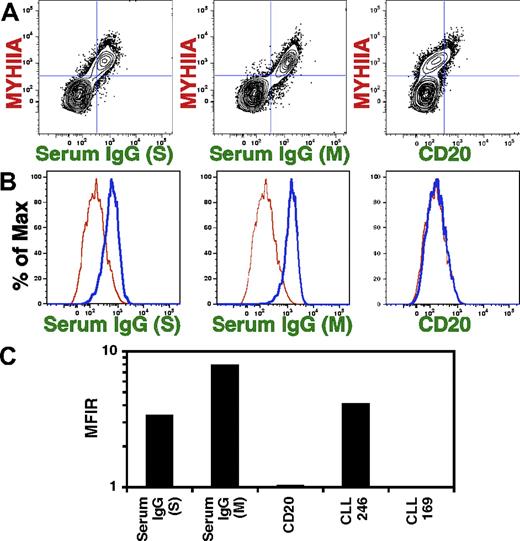
Binding to apoptotic cells is a characteristic of human serum natural antibodies.17,18 Therefore, we tested human serum for binding to MEACs (Figure 5). To match the isotype of our recombinant CLL mAbs, we tested IgG purified from human serum from 2 different vendors. Both sources of serum IgG bound MEACs (Figure 5A; left and middle panels). Natural antibodies, which consist of IgM, IgG, and IgA molecules, are produced without overt antigen selection.30 In comparison, a human IgG1 antibody, which had been selected for specificity to the CD20 antigen, did not bind MEACs (Figure 5A right panel). After gating on MYHIIA+ apoptotic cells, histograms clearly show serum IgG binding to MEACs (Figure 5B left and middle panels) with a higher MFI than that of CLL 169, a mutated CLL mAb with only background binding to MEACs (Figure 3). MEAC binding of human IgG anti-CD20 mAb is similar to that of CLL 169 (Figure 5B right panel), suggesting that mutated or overt antigen-selected antibodies do not have the same binding characteristics as natural antibodies. Serum IgG and anti-CD20 mAb binding to MEACs was quantified by MFIR (Figure 5C), reflecting the previous observations (Figure 5A-B). In addition, these MFIR measurements show that natural antibody binding to MEACs is similar to that for a high MEAC binding CLL mAb (CLL 246; Figure 5C). Thus, because polyclonal serum natural antibodies react to MEACs, at least a subset of CLL leukemic clones may derive from natural antibody producing B lymphocytes.
Normal human serum IgG reacts with MEACs. Spontaneous apoptotic Jurkat cells were stained with 25 μg/mL rabbit anti–human MYHIIA antibody and 25 μg/mL CLL mAb, normal human serum IgG from Sigma (S; 25 μg/mL) or Miltenyi Biotec (M; 2 μL), or 25 μg/mL humanized anti-CD20 mAb, followed by 1 μg/mL secondary antibodies: PE-conjugated anti–rabbit IgG and FITC-conjugated anti–human IgG. Representatives of 4 experiments are shown. (A) Flow cytometric analyses are displayed as contour plots of fluorescence intensity shown on log scales with BiExponential transformation. Panels show costaining with anti-MYHIIA and serum IgG or anti-CD20. (B) After gating on MYHIIA+ cells, histograms of the percentage of maximum (% of Max) fluorescent intensity for indicated staining (thick blue line) is shown relative to cells stained with CLL 169 mAb (thin red line). (C) The MFIR staining of MEACs was calculated as in Figure 3C except that the MFIR was calculated by dividing the MFI for MYHIIA+ cells stained with CLL 169 mAb.
Normal human serum IgG reacts with MEACs. Spontaneous apoptotic Jurkat cells were stained with 25 μg/mL rabbit anti–human MYHIIA antibody and 25 μg/mL CLL mAb, normal human serum IgG from Sigma (S; 25 μg/mL) or Miltenyi Biotec (M; 2 μL), or 25 μg/mL humanized anti-CD20 mAb, followed by 1 μg/mL secondary antibodies: PE-conjugated anti–rabbit IgG and FITC-conjugated anti–human IgG. Representatives of 4 experiments are shown. (A) Flow cytometric analyses are displayed as contour plots of fluorescence intensity shown on log scales with BiExponential transformation. Panels show costaining with anti-MYHIIA and serum IgG or anti-CD20. (B) After gating on MYHIIA+ cells, histograms of the percentage of maximum (% of Max) fluorescent intensity for indicated staining (thick blue line) is shown relative to cells stained with CLL 169 mAb (thin red line). (C) The MFIR staining of MEACs was calculated as in Figure 3C except that the MFIR was calculated by dividing the MFI for MYHIIA+ cells stained with CLL 169 mAb.
Discussion
We have found that MYHIIA is exposed on a subgroup of apoptotic cells (Figures 1–2), which could represent one way that CLL subset 6 BCRs interact with MYHIIA autoantigen in vivo. In support of this idea, we found that CLL subset 6 mAbs in vitro recognize MEACs, and not apoptotic cells without exposed MYHIIA or live cells (Figures 1,Figure 2–3). Thus, these results confirm our previous observations that CLL subset 6 mAbs recognize MYHIIA7 and further suggest that exposure to MEACs could stimulate the BCRs of leukemic clones from this subset. In CLL, MEACs could come from CLL cell turnover (average, 0.5%-2.3% deaths per day31,32 ), normal cell turnover, or induction of damage in vivo (eg, ischemia reperfusion injury induces exposure of MYHIIA33 ). In our studies, we produced MEACs from Jurkat T cells undergoing spontaneous apoptosis. However, we have also observed MEAC formation in a human B-cell line (RAMOS) and in CLL cells undergoing apoptosis in vitro (C.C.C., N.C., B.M.A., and L.Z., unpublished data, November 2008-January 2009). Thus, MEACs form when multiple cell types undergo apoptosis; we are currently investigating the requirements for MEAC production.
Because many CLL mAbs bind apoptotic cells,13,14 we tested a panel of 26 CLL mAbs for reactivity to the MEAC fraction of cells undergoing apoptosis (Figure 3; Table 1). More than 60% of CLL mAbs tested bound well to MEACs (MFIR ≥ 1.5); this included 14 mAbs that did not belong to subset 6. This percentage of reactive CLL mAbs corresponds with our previous observation of apoptotic cell binding,13 with a correspondence in all CLL mAbs except for 3 patients (CLL 141, 258, 342). CLL 258 mAb was negative for binding to apoptotic Jurkat and RAMOS cells,13 whereas in this study, CLL 258 mAb bound MEACs well (MFIR = 2.1). For CLL 141 and 342, we observed low MEAC binding (MFIR = 0.7 and 1.1, respectively), whereas these 2 CLL mAbs were positive for binding to apoptotic Jurkat and RAMOS cells in the prior study.13 In addition, the percentage of apoptotic cell binding in the prior study13 sometimes differed in relative degree compared with the MEAC binding level. For example, in the prior study, CLL 114 and 845 had relatively modest percentage apoptotic cell binding (“+”),13 whereas in this study, using a different type of assay, we observed a relatively high level of MEAC binding (MFIR = 17.05 and 25.25, respectively). Conversely, in the prior study, CLL 014 had a very high percentage of apoptotic cell binding (“+++”),13 whereas the level of MEAC binding was relatively not as high (MFIR = 1.84). These differences in absolute and relative reactivity to apoptotic cells can be explained in 2 ways. First, in the cases where the prior study had no (CLL 258) or relatively little (CLL 114 and 845) percentage of apoptotic cell binding compared with higher MEAC binding in this study, these differences may be explained by a low level of MEAC induction in the prior study. Second, in the cases where the analyses used in this study indicated low (CLL 141 and 342) or not as high (CLL 014) MEAC binding compared with a higher percentage apoptotic cell binding in the prior study, these differences may be explained by reactivity to non-MEAC epitopes. For example, CLL141 and 342 mAbs are encoded by IGHV4-34 gene segments, which can react to the N-acetyllactosamine carbohydrate of the I/i blood group antigen.34 This study restricted measurements to just the MEAC subset.
It is important to recognize that reactivity of other non–subset 6 CLL mAbs to MEACs does not necessarily indicate reactivity to MYHIIA. Indeed, this binding likely reflects reactivity to other autoantigens, such as, but not restricted to, vimentin, filamin B, or oxidized epitopes,13,14 that are induced during apoptosis in the MEAC fraction. At present, the specific autoantigen reactivities of these other non–subset 6 CLL mAbs are not clear. Preliminary studies of subset 8 (CLL 114) and subset 9 (CLL 246 and CLL 355) mAbs showed a predominant cytoplasmic staining pattern on nonpermeabilized apoptotic human Jurkat T cells (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) similar to that observed for permeabilized human HEp-2 epidermal cells.13,22 Costaining of apoptotic Jurkat cells with anti-MYHIIA revealed that these subset 8 and 9 CLL mAbs bound primarily MEACs (supplemental Figure 1), which was confirmed quantitatively by flow cytometry (Figure 3). Examination of colocalization of subset 8 and 9 CLL mAbs with anti-MYHIIA staining by color overlap suggested that MYHIIA colocalizes to areas of CLL mAb staining, but that these CLL mAbs also bind to areas distinct from those bound by anti-MYHIIA antibodies. Intensity correlation analyses affirmed this observation by enumerating the CLL mAb and anti-MYHIIA colocalization with r values (r = .418, .521, and .558 for CLL 114, 246, and 355, respectively); these correlations were slightly less than that observed of CLL 068 mAb (r = .647; Figure 1C). Therefore, subset 8 and 9 CLL mAbs either react to multiple antigens including MYHIIA, or to an antigen that can be associated with MYIIIA. In support of the latter idea, the r values for F-actin colocalization with MYHIIA in SNB19 human glioblastoma or HEp-2 cells7 were similar to those observed for subset 8 and 9 CLL mAbs. Incomplete colocalization of MYHIIA and F-actin is expected, because MYHIIA binds reversibly to F-actin to form transient intracellular motor complexes involved in cellular movement and morphology.11 In addition, we have found in a prior study that the subset 8 CLL 114 mAb binds vimentin,7 although CLL 114 mAb may bind other (auto)antigen(s), and the antigen(s) that are bound by subset 9 CLL mAbs remain to be elucidated. The identification of the specific antigens that bind to non–subset 6 CLL mAbs is beyond the scope of this paper and is currently under study.
Of note, CLL mAbs from the same stereotyped subsets bound with similar effectiveness to MEACs (Figure 3C; Table 1), perhaps validating the hypotheses that members of stereotyped subsets bind similar epitopes,4,8,24,35 and that these groupings have functional utility. Subsets 1, 6, 8, 9, and 28 with predominantly unmutated IGHV CLL mAbs generally bound well (MFIR ≥ 1.5), whereas subsets 2 and 4, usually characterized by mutated IGHV, did not bind well to MEACs (MFIR < 1.5). Subset 8 CLL mAbs demonstrated uniformly high levels of MEAC binding activity, possibly due to multiple factors, such as higher-affinity interaction with an uncharacterized molecule exposed on MEACs and/or reactivity with multiple uncharacterized antigens exposed on MEACs. We have noted that a CLL mAb from this subset, characterized by an unmutated IGHV4-39, IGHD6-13, and IGHJ5 gene rearrangement, can bind vimentin,7 a molecule reported to be exposed during apoptosis.16 In contrast, subset 9 CLL mAbs exhibited a wide range of MEAC binding levels (MFIR from 1.84 to 22.55), possibly reflecting diverse HCDR3 sequences (Table 1). This diversity may permit dissection of the amino acid sequence requirements for MEAC binding.
CLL mAb binding to MEACs inversely correlated with the level of IGHV mutation (Figure 4A), suggesting that MEAC binding may predict patient survival. Available clinical data for 24 of the 26 patients with CLL from whom the tested mAbs were derived showed a remarkable correlation between high MFIR binding and poor patient survival (Figure 4B; Tables 1–2). This correlation was statistically significant, whereas the correlation of patient survival with levels of IGHV mutation, CD38, or ZAP-70 in the same patients did not reach significance (Figure 4C; Tables 1–2). The difference in significance between MEAC binding and IGHV mutation was due to one patient with mutated (CLL 154) IGHV CLL and 2 patients with unmutated (CLL 376 and 412) IGHV CLL having survival outcomes contrary to that expected for their IGHV mutation status, using the generally used 2.0% cutoff threshold for mutation status. Of these 3 patients, 2 (CLL 154 and 412) have borderline IGHV mutation status (2.1% and 1.8%, respectively). Interestingly, CLL 154 has only 3 replacement mutations in the IGHV protein, all of which are in framework regions. Because the HCDRs are unaffected in CLL 154, antigen binding could be similar to other subset 1 CLL mAbs, perhaps further indicating why this borderline IGHV-mutated case incorrectly predicts antigen-binding ability and survival. In contrast, CLL 412, which has an unmutated IGHV, has 2 replacement mutations in the HCDR. Thus, CLL 412 mAb with borderline unmutated IGHV may have sufficient changes in its HCDR to alter antigen-binding specificity, explaining its behavior as a “mutated” CLL mAb. Although such discrepancies in the prognostic implications of IGHV mutation status and MEAC binding may support modifying the IGHV mutation status threshold, as some have proposed,36-38 we believe that these discrepancies may highlight the relative importance of a true functional antigen-binding parameter (as determined by MEAC reactivity) versus presumed antigen-binding site structure (as gleaned from IGHV mutation status). Although this intriguing possibility will require confirmation in a larger number of patients, we favor this possibility because it links directly BCR structure with clinical course and supports the role of antigen binding to the development and evolution of the disease.5-7
CLL subset 4 mAbs are of the IgG isotype and have mutated rearrangements involving IGHV4-34.4,8 Patients with these mAbs generally have good survival outcomes,34 in agreement with our MEAC-binding results (Tables 1–2). Based on one model of CLL development,5-7 diminished reactivity with MEACs would result in the lack of BCR stimulation of the leukemic cell, leading to impaired survival and less growth of the CLL clone and therefore better patient outcome. However, the reactivity of this subset of CLL mAbs with other antigens, such as the carbohydrate I/i blood group antigen and possibly Epstein-Barr virus or cytomegalovirus antigens,34 suggests that the explanation for better patient survival is complex. Interestingly, all 3 tested CLL subset 4 mAbs retain all the hydrophobic patch residues involved in binding to the I/i antigen;39 perhaps binding to this carbohydrate antigen does not lead to expansion, but instead results in anergy of the leukemic clone.40,41
Binding to apoptotic cells is a property of natural antibodies.17,18 Therefore, we tested human serum antibodies from healthy persons for the ability to bind MEACs. Polyclonal human serum IgG from 2 different sources bound MEACs well and at a level comparable with high MEAC-binding CLL mAbs (Figure 5). Because natural antibodies only make up a fraction of polyclonal serum IgG, the direct comparison of MEAC binding levels to CLL mAbs is difficult to ascertain clearly. However, these data imply that a large number of natural antibodies bind epitopes revealed on MEACs and/or a small number of natural antibodies bind MEACs with high efficiency. Interestingly, serum IgG only bound MYHIIA+ cells, with very little detectable binding to MYHIIA− cells, suggesting that binding of natural antibodies to apoptotic cells may be restricted to the MEAC subgroup. This reactivity may represent specificity for MYHIIA, as natural antibody reactivity to MYHIIA has been described;33,42 however, because natural antibodies are composed of immunoglobulins with multiple specificities,43 other antigenic determinants may also be exposed on MEACs. We speculate that exposure during apoptosis of vimentin, filamin B, and other antigens generated by chemical modifications are not only made available to some CLL cells,7,13,14 but also to subsets of normal B-lymphocytes producing natural antibodies, thereby linking these leukemic and normal B cells to the MEAC subgroup through common antigenic reactivities.
Thus, our data provide evidence that many CLL mAbs of stereotyped and nonstereotyped subsets, as well as a fraction of natural antibodies, recognize antigenic targets on MEACs. This in turn suggests that at least a large subset of CLL mAbs derive from B cells that produce natural antibodies. These B cells could be derived from the human counterparts of mouse B-1 cells or other subsets producing natural antibodies.17 Some of these cells could be intermittently exposed to and stimulated by MEACs, leading to the development and/or expansion of CLL, ultimately leading to poor patient outcome. Finally, our finding that antigen (eg, MEAC) binding, in particular by members of stereotyped subsets, may better indicate patient outcome than IGHV mutation status is provocative and requires further investigation.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors appreciate the assistance of Barbara Napolitano and Nina Kohn (Biostatistics Unit, The Feinstein Institute for Medical Research) for statistical analysis, Yayu Chuang (The Feinstein Institute for Medical Research) for immunofluorescence advice, Stella Stefanova (Flow Cytometry Facility, The Feinstein Institute for Medical Research) for flow cytometry, and Klaus Dittmar, Dale M. Janson, Bill Kennedy, Jonathan D. Weinberger, and Chris E. Thomas for collection of clinical data. We also thank Bettie M. Steinberg, Manuela Woelfle, Patricia Mongini, and Sophia Yancopoulos (The Feinstein Institute for Medical Research) for helpful discussions regarding manuscript preparation.
This work was supported by an R01 grant from the National Institutes of Health (NIH; CA81554 to N.C.), an M01 General Clinical Research Center grant from the NIH (RR01853 to N.C.), The Karches Foundation, The Prince Family Foundation, The Marks Foundation, The Jerome Levy Foundation, The Leon Levy Foundation, the Tebil Foundation Inc, and the Joseph Eletto Leukemia Research Fund.
National Institutes of Health
Authorship
Contribution: C.C.C., R.C., L.Z., R.N.D., and N.C. designed research and analyzed results; R.C., L.Z., S.D., B.M.A, and R.N.D. performed experiments; C.C.C. and N.C. wrote the paper and made the figures; and M.S.K., J.E.K., S.L.A., and K.R.R. contributed clinical data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Charles C. Chu, The Feinstein Institute for Medical Research, 350 Community Dr, Manhasset, NY 11030; e-mail: cchu@nshs.edu.