Abstract
The importance of donor-recipient human leukocyte antigen (HLA)-DPB1 matching for the clinical outcome of unrelated hematopoietic stem cell transplantation (HSCT) is controversial. We have previously described an algorithm for nonpermissive HLA-DPB1 disparities involving HLA-DPB1*0901,*1001,*1701,*0301,*1401,*4501, based on T-cell alloreactivity patterns. By revisiting the immunogenicity of HLA-DPB1*02, a modified algorithm was developed and retrospectively tested in 621 unrelated HSCTs facilitated through the Italian Registry for oncohematologic adult patients. The modified algorithm proved to be markedly more predictive of outcome than the original one, with significantly higher Kaplan-Meier probabilities of 2-year survival in permissive compared with nonpermissive transplantations (55% vs 39%, P = .005). This was the result of increased adjusted hazards of nonrelapse mortality (hazard ratio [HR] = 1.74; confidence interval [CI], 1.19-2.53; P = .004) but not of relapse (HR = 1.02; CI, 0.73-1.42; P = .92). The increase in the hazards of overall mortality by nonpermissive HLA-DPB1 disparity was similar in 10 of 10 (HR = 2.12; CI, 1.23-3.64; P = .006) and 9 of 10 allele-matched transplantations (HR = 2.21; CI, 1.28-3.80; P = .004), both in early-stage and in advanced-stage disease. These data call for revisiting current HLA matching strategies for unrelated HSCT, suggesting that searches should be directed up-front toward identification of HLA-DPB1 permissive, 10 of 10 or 9 of 10 matched donors.
Introduction
Human leukocyte antigen (HLA)-DP was first described in 1980 as a distinct group of HLA class II antigens eliciting variable T-cell responses in mixed lymphocyte reactions (MLRs).1 Since then, increasing evidence has accumulated to show that HLA-DP molecules function as bona fide restriction elements for viral and tumor antigen-specific T cells2-4 and can elicit both humoral and cellular alloresponses relevant in clinical transplantation.5-7 The HLA-DP antigens are αβ heterodimers encoded by the genes of the DPA1 locus, which displays limited polymorphism, and of the highly polymorphic DPB1 locus, with 132 alleles coding for 116 different proteins described to date (http://www.ebi.ac.uk./imgt/hla). Because of weak linkage disequilibrium with other HLA class II loci,8 unrelated hematopoietic stem cell transplantation (HSCT) is generally performed across allelic HLA-DPB1 mismatches. The definition of nonpermissive mismatches for this locus has therefore important practical implications. Based on cross-reactive T-cell alloreactivity patterns, our group has previously described an algorithm for nonpermissive HLA-DPB1 mismatches, which were shown by us and subsequently by others to be significantly associated with transplant-related mortality but not with overall mortality (OM).9-11
In the present study, we have modified this algorithm by integrating our functional data with those reported by others12-14 and tested its clinical predictive value by retrospective analysis of 621 unrelated HSCT facilitated through the Italian Bone Marrow Donor Registry. The results provide compelling evidence that the clinical outcome of unrelated HSCT can be significantly improved by consideration of nonpermissive HLA-DPB1 disparity in donor selection.
Methods
Patients
A total of 621 adult oncohematologic patients (>18 years) received an HSCT from an unrelated donor (UD) in Italy between 1999 and 2006, after informed consent was obtained in accordance with the Declaration of Helsinki for protocols approved by the ethical committees of the different participating centers. Eligible diagnoses are listed in Table 1. Clinical data were obtained from Gruppo Italiano Trapianto di Midollo Osseo, CSE e Terapia Cellulare registry, whereas the HLA typing results were collected through the Italian Bone Marrow Donor Registry.15 According to disease status at HSCT, patients were categorized into 3 disease groups: CML in first chronic phase (CML-CP1; n = 31), acute leukemia in first complete remission (AL-CR1; n = 127), and all other diseases including advanced-stage leukemia (ADV; n = 463). HSCT conditioning regimens were either myeloablative (MA) or reduced intensity (RIC), according to local or cooperative protocols (Table 1). Graft-versus-host disease (GVHD) prophylaxis was mostly performed with cyclosporine and methotrexate.
Patient, donor, and transplantation characteristics
| Variable . | n . |
|---|---|
| Donor sex, male/female | 438/183 |
| Donor median age, y (range) | 35 (19-56) |
| Donor CMV status, positive/negative/missing | 327/284/10 |
| Patient sex, male/female | 352/269 |
| Patient median age, y (range) | 41 (18-66) |
| Patient CMV status, positive/negative/missing | 336/77/208 |
| HSCT year | |
| 1999-2000 | 13/35 |
| 2001-2002 | 47/84 |
| 2003-2004 | 113/111 |
| 2005-2006 | 141/77 |
| ATG use, yes/no/missing | 422/153/46 |
| Diagnosis | |
| Acute lymphoblastic leukemia | 119 |
| Acute myeloid leukemia | 193 |
| Chronic lymphocytic leukemia | 11 |
| Chronic myeloid leukemia | 76 |
| Hodgkin lymphoma | 24 |
| Myelodysplastic syndrome | 63 |
| Multiple myeloma | 41 |
| Myeloproliferative syndrome | 21 |
| Non-Hodgkin lymphoma | 60 |
| Secondary acute leukemia | 13 |
| Disease group, CML-CP1/AL-CR1/ADV | 31/127/463 |
| Stem cell source, PBSC/BM/missing | 249/368/4 |
| Conditioning, MA/RIC/missing | 427/161/33 |
| TBI use, yes/no/missing | 359/250/12 |
| HLA matching | |
| 12 of 12 allele-matched | 41 |
| 10 of 10 allele-matched, DPB1 mismatched | 201 |
| 9 of 10 allele-matched, DPB1 mismatched | 199 |
| Less than or equal to 8 of 10 allele-matched, DPB1 mismatched | 137 |
| DPB1-matched, mismatched at other loci | 43 |
| DRB3/4/5-matched/DRB3/4/5 typed | 550/616 |
| DQA1-matched/DQA1 typed | 516/537 |
| DPA1-matched/DPA1 typed | 146/277 |
| Permissive/nonpermissive HLA-DPB1 disparity | |
| Mismatched permissive TCE3/TCE4 | 287/158 |
| Mismatched nonpermissive (GvH) TCE3/TCE4 | 145/219 |
| Mismatched nonpermissive (HvG) TCE3/TCE4 | 105/160 |
| Variable . | n . |
|---|---|
| Donor sex, male/female | 438/183 |
| Donor median age, y (range) | 35 (19-56) |
| Donor CMV status, positive/negative/missing | 327/284/10 |
| Patient sex, male/female | 352/269 |
| Patient median age, y (range) | 41 (18-66) |
| Patient CMV status, positive/negative/missing | 336/77/208 |
| HSCT year | |
| 1999-2000 | 13/35 |
| 2001-2002 | 47/84 |
| 2003-2004 | 113/111 |
| 2005-2006 | 141/77 |
| ATG use, yes/no/missing | 422/153/46 |
| Diagnosis | |
| Acute lymphoblastic leukemia | 119 |
| Acute myeloid leukemia | 193 |
| Chronic lymphocytic leukemia | 11 |
| Chronic myeloid leukemia | 76 |
| Hodgkin lymphoma | 24 |
| Myelodysplastic syndrome | 63 |
| Multiple myeloma | 41 |
| Myeloproliferative syndrome | 21 |
| Non-Hodgkin lymphoma | 60 |
| Secondary acute leukemia | 13 |
| Disease group, CML-CP1/AL-CR1/ADV | 31/127/463 |
| Stem cell source, PBSC/BM/missing | 249/368/4 |
| Conditioning, MA/RIC/missing | 427/161/33 |
| TBI use, yes/no/missing | 359/250/12 |
| HLA matching | |
| 12 of 12 allele-matched | 41 |
| 10 of 10 allele-matched, DPB1 mismatched | 201 |
| 9 of 10 allele-matched, DPB1 mismatched | 199 |
| Less than or equal to 8 of 10 allele-matched, DPB1 mismatched | 137 |
| DPB1-matched, mismatched at other loci | 43 |
| DRB3/4/5-matched/DRB3/4/5 typed | 550/616 |
| DQA1-matched/DQA1 typed | 516/537 |
| DPA1-matched/DPA1 typed | 146/277 |
| Permissive/nonpermissive HLA-DPB1 disparity | |
| Mismatched permissive TCE3/TCE4 | 287/158 |
| Mismatched nonpermissive (GvH) TCE3/TCE4 | 145/219 |
| Mismatched nonpermissive (HvG) TCE3/TCE4 | 105/160 |
HLA typing
All 621 pairs were typed for HLA-A, B, C, DRB1, DQB1, DPB1, and 616, 537, and 277 pairs also for DRB3/4/5, DQA1, and DPA1, respectively, by standard methods, including sequence-specific oligonucleotide probing, sequence-specific priming, and/or sequence based typing. For all loci tested, typing was performed to the 4-digit level, according to the quality standards of the European Federation of Immunogenetics, which foresees resolution of all alleles differing for exons 2 and 3 for HLA class I, and exon 2 for HLA class II, as well as all Null alleles (http://www.efiweb.eu/index.php?id = 102).
Clinical endpoint definitions
Overall survival (OS), nonrelapse mortality (NRM), graft failure, and relapse incidence (RI) were defined according to European Group for Blood and Marrow Transplantation criteria (http://www.ebmt.org/). Grading of acute GVHD (aGVHD) was performed according to current criteria.16
Statistical analysis
Continuous variables were expressed as median (range), whereas categorical ones were expressed as proportions. χ2 and Mann-Whitney U tests were used for comparisons between major clinical parameters in permissive and nonpermissive pairs for categorical and continuous variables, respectively, and did not reveal any significant differences between the 2 groups. Probabilities of OS with respective 95% confidence interval (CI) were calculated using the Kaplan-Meier estimator,17 and survival curves were compared using the log-rank test.18 Univariate regression analysis was used to test association between HLA or non-HLA variables and OM, NRM, RI (Cox regression),19 aGVHD, and graft failure (logistic regression). For numerical reasons, in univariate and multivariate analyses, pairs with more than or equal to 2 HLA mismatches at loci other than HLA-DPB1 were grouped together. Variables with a P value less than .2 were included in the multivariate analysis, and only variables with a P value less than .05 were retained in the final multivariate model. Non-HLA factors included donor sex, age (continuous variable), and CMV status; patient sex, age (continuous variable), CMV status; year of transplantation (continuous variable), use of antithymocyte globulin as GVHD prophylaxis, disease group (CML-CP1 vs AL-CR1 vs ADV), stem cell source (peripheral blood vs bone marrow), conditioning regimen (MA vs RIC), use of total body irradiation. HLA-DPB1 permissiveness was tested for interaction with the number of HLA mismatches, and the term of interaction was 0.03 for both OM and NRM (0-1 vs >1 HLA mismatch). No significant interactions resulted with regards to the other clinical endpoints.
Results
A modified algorithm for nonpermissive HLA-DPB1 mismatches
We have previously described an algorithm for nonpermissive HLA-DPB1 disparities, on the basis of cross-reactive patterns by alloreactive T cells involved in HSCT rejection targeted to HLA-DPB1*0901.11 This algorithm foresees group-specific rather than allele-specific HLA-DPB1 matching, dividing HLA-DPB1 alleles into 3 groups with high (HLA-DPB1*0901,*1001,*1701), intermediate (HLA-DPB1 *0301,*1401,*4501), or low (most other HLA-DPB1 alleles) immunogenicity,11,20 presumably on the basis of a shared alloreactive T-cell epitope (TCE). The modality of classification of donor-recipient pairs as permissive or nonpermissive according to this 3-group algorithm, henceforward referred to as TCE3, is described in Figure 1.11,13,14,20,21 By retrospective analysis of unrelated HSCT stratified according to TCE3, we and others showed an association of nonpermissive HLA-DPB1 mismatches with transplant-related mortality, grade 2 to 4 aGVHD, and graft rejection, in 10 of 10 matched pairs.9-11,22
An algorithm for nonpermissive HLA-DPB1 disparities according to TCE3 or TCE4. (A) HLA-DPB1 alleles were classified into 3 groups (TCE3), or 4 groups (TCE4), on the basis of T-cell alloreactivity. TCE3 group 1 and TCE4 group 1: Alleles encoding antigens recognized by all T-cell clones studied by Zino et al.11 TCE3 group 2 and TCE4 group 2: Alleles encoding antigens recognized by some but not all T-cell clones studied by Zino et al.11 TCE 3 group 3: Alleles encoding antigens recognized by none of the T-cell clones studied by Zino et al.11 “Others” refers to all alleles that can be classified according to the algorithm of Zino et al.20 TCE4 group 3: DPB1*02, encoding antigens eliciting intermediate levels of MLR reactivity.13,14,21 TCE4 group 4: All alleles from TCE3 group 3 except for DPB1*02. (B) The 3 or 4 groups of HLA-DPB1 alleles can be present in different combinations in diploid cells. Numbers indicate the group of the first (before the slash) and the second (after the slash) HLA-DPB1 allele of donor or recipient. Classification of HLA-DPB1 group disparities as permissive or nonpermissive in GvH or HvG direction is indicated for all possible combinations. Note that all nonpermissive TCE3 disparities are also TCE4-nonpermissive (gray boxes). In contrast, only a part of the TCE3-permissive disparities are permissive also according to TCE4 (white boxes), whereas the remaining TCE3-permissive disparities score as nonpermissive in TCE4 (striped boxes).
An algorithm for nonpermissive HLA-DPB1 disparities according to TCE3 or TCE4. (A) HLA-DPB1 alleles were classified into 3 groups (TCE3), or 4 groups (TCE4), on the basis of T-cell alloreactivity. TCE3 group 1 and TCE4 group 1: Alleles encoding antigens recognized by all T-cell clones studied by Zino et al.11 TCE3 group 2 and TCE4 group 2: Alleles encoding antigens recognized by some but not all T-cell clones studied by Zino et al.11 TCE 3 group 3: Alleles encoding antigens recognized by none of the T-cell clones studied by Zino et al.11 “Others” refers to all alleles that can be classified according to the algorithm of Zino et al.20 TCE4 group 3: DPB1*02, encoding antigens eliciting intermediate levels of MLR reactivity.13,14,21 TCE4 group 4: All alleles from TCE3 group 3 except for DPB1*02. (B) The 3 or 4 groups of HLA-DPB1 alleles can be present in different combinations in diploid cells. Numbers indicate the group of the first (before the slash) and the second (after the slash) HLA-DPB1 allele of donor or recipient. Classification of HLA-DPB1 group disparities as permissive or nonpermissive in GvH or HvG direction is indicated for all possible combinations. Note that all nonpermissive TCE3 disparities are also TCE4-nonpermissive (gray boxes). In contrast, only a part of the TCE3-permissive disparities are permissive also according to TCE4 (white boxes), whereas the remaining TCE3-permissive disparities score as nonpermissive in TCE4 (striped boxes).
The patient from whom the T-cell clones tested to define the TCE3 were derived shared HLA-DPB1*0201 with her stem cell donor.23 Because of negative selection of potentially self-reactive T cells from the patient's repertoire, the T-cell clones used for establishing the algorithm were not informative for HLA-DPB1*02, which might encode a second, distinct immunogenic TCE. The existence of such an epitope is indeed suggested by previous reports showing that HLA-DBP1*0201 elicits T-cell responses in classic MLR, although apparently to a lower extent compared with the antigens encoded by the immunogenic alleles from the 3-group algorithm.13,14,21 On the basis of these observations, we designed a 4-group algorithm, including HLA-DPB1*02 as a separate group with immunogenicity lower than that of group 2 alleles but higher than that of the low immunogenic alleles. This algorithm is henceforward referred to as TCE4 and is described in Figure 1.
HLA matching of patients and donors
Of the 621 pairs studied, 41 were 4-digit matched for all 12 HLA alleles, including DPB1 (12 of 12), whereas 43 were identical for both HLA-DPB1 alleles but presented one or more mismatches at other HLA loci, for a total of 84 DPB1 matched pairs. The remaining 537 pairs presented at least one allelic mismatch at HLA-DPB1, with zero (10 of 10; n = 201), one (9 of 10; n = 199), or more than one (≤8 of 10; n = 137) allelic or antigenic mismatches at the other loci. A total of 616 pairs were also typed for HLA-DRB3/4/5; of these, only 66 pairs (10.7%) had a mismatch for at least one of these 3 loci, whereas the remaining 550 pairs (89.3%) were matched (Table 1). A total of 537 and 277 pairs were also typed for HLA-DQA1 and DPA1, respectively. Only 21 of 537 pairs (3.9%) were DQA1 mismatched, whereas 131 of 277 pairs (47.3%) presented mismatches at DPA1 (Table 1).
On the basis of HLA-DPB1 frequencies in the white population,20 it can be predicted that permissive HLA-DPB1 mismatches according to TCE3 or TCE4 are present in approximately 55% and 30% of pairs, respectively. This was confirmed in our cohort of 537 HLA-DPB1 allele-mismatched pairs. When classified according to TCE3, 287 (53.4%) scored as permissive, and 145 (27.0%) or 105 (19.6%) as nonpermissive in graft-versus-host (GvH) or host-versus-graft (HvG) direction, respectively. When classified according to TCE4, 158 of 537 (29.4%) scored as permissive and 219 of 537 (40.8%) or 160 of 537 (29.7%) as nonpermissive in GvH or HvG direction, respectively (Table 1).
Clinical outcome
Kaplan-Meier estimates of survival.
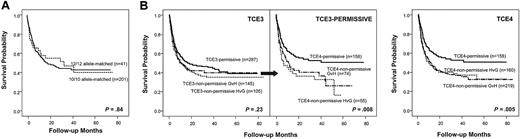
The observed probability of survival at 2 years in the total cohort was 44% (273 of 621 patients). In line with previous reports,24,25 observed survival probabilities in these 621 patients were not markedly different after HLA-DPB1 allele-matched (43%) versus mismatched (44%) transplantations. In addition, when 242 transplantations fully matched for HLA-A, B, C, DRB1, and DQB1 alleles were considered separately, allelic DPB1 mismatches did not have a significant impact on the probability of OS (95% CI) at 2 years, which was 55% (39%-70%) and 47% (39%-54%) for DPB1 allele-matched (12 of 12, n = 41) or mismatched (10 of 10, n = 201) transplantations, respectively (P = .84; Figure 2A). When pairs with allelic DPB1 disparities were further subdivided into those with permissive (n = 287) or nonpermissive GvH (n = 145) or HvG (n = 105) disparities according to TCE3, there was a trend for worse survival in the nonpermissive pairs which, however, was not statistically significant (47% [41%-53%] in the permissive vs 38% [30%-46%] in the nonpermissive GvH and 41% [31%-51%] in the nonpermissive HvG group; P = .23; Figure 2B left panel). This is in line with previous observations made by us and others, which failed to document a significant association between nonpermissive DPB1 disparities according to TCE3, and OS in global cohorts of patients with various degrees of matching for the other loci.9-11 TCE3-permissive pairs could be further subdivided into those classified as permissive also according to TCE4 (n = 158), and those classified as nonpermissive in GvH (n = 74) or HvG (n = 55) direction according to TCE4. Interestingly, the 2-year OS probabilities associated with these nonpermissive TCE4 disparities were 40% (28%-52%) for the GvH and 36% (23%-49%) for the HvG group, significantly lower compared with the permissive TCE4 disparities (55% [46%-63%]; P = .008; Figure 2B middle panel). This translated into a significant impact of nonpermissive HLA-DPB1 TCE4 disparities on OS in the total cohort, with 2-year survival probabilities of 55% (46%-63%), 39% (32%-45%), and 40% (32%-47%) in the permissive (n = 155), GvH (n = 219), or HvG (n = 160) group, respectively (P = .005; Figure 2B right panel). Given the similar impact of nonpermissive GvH and HvG mismatches on survival (P = .94 in the heterogeneity test) and all other clinical endpoints studied (data not shown), data for the 2 groups from here on are shown together.
Impact of allelic or allele-group HLA-DPB1 disparities on OS after unrelated HSCT. Shown are Kaplan-Meier estimates of survival. (A) HLA-A, B, C, DRB1, and DQB1 matched transplantations (n = 242), stratified according to the presence (10 of 10; —) or absence (12 of 12;  ) of allelic DPB1 mismatches. (B) Left panel: All HLA-DPB1 mismatched transplantations (n = 537), stratified according to the presence of TCE3-permissive (—) or TCE3-nonpermissive HvG (
) of allelic DPB1 mismatches. (B) Left panel: All HLA-DPB1 mismatched transplantations (n = 537), stratified according to the presence of TCE3-permissive (—) or TCE3-nonpermissive HvG ( ) or GvH (dash-dot line) mismatches. Middle panel: TCE3-permissive transplantations (n = 287), subdivided into those permissive also according to TCE4 (—), or those TCE4-nonpermissive in HvG (
) or GvH (dash-dot line) mismatches. Middle panel: TCE3-permissive transplantations (n = 287), subdivided into those permissive also according to TCE4 (—), or those TCE4-nonpermissive in HvG ( ) or GvH (dash-dot line). Right panel: All HLA-DPB1 mismatched transplantations (n = 537), stratified according to the presence of TCE4-permissive (—) or TCE4-nonpermissive HvG (
) or GvH (dash-dot line). Right panel: All HLA-DPB1 mismatched transplantations (n = 537), stratified according to the presence of TCE4-permissive (—) or TCE4-nonpermissive HvG ( ) or GvH (dash-dot line) mismatches.
) or GvH (dash-dot line) mismatches.
Impact of allelic or allele-group HLA-DPB1 disparities on OS after unrelated HSCT. Shown are Kaplan-Meier estimates of survival. (A) HLA-A, B, C, DRB1, and DQB1 matched transplantations (n = 242), stratified according to the presence (10 of 10; —) or absence (12 of 12;  ) of allelic DPB1 mismatches. (B) Left panel: All HLA-DPB1 mismatched transplantations (n = 537), stratified according to the presence of TCE3-permissive (—) or TCE3-nonpermissive HvG (
) of allelic DPB1 mismatches. (B) Left panel: All HLA-DPB1 mismatched transplantations (n = 537), stratified according to the presence of TCE3-permissive (—) or TCE3-nonpermissive HvG ( ) or GvH (dash-dot line) mismatches. Middle panel: TCE3-permissive transplantations (n = 287), subdivided into those permissive also according to TCE4 (—), or those TCE4-nonpermissive in HvG (
) or GvH (dash-dot line) mismatches. Middle panel: TCE3-permissive transplantations (n = 287), subdivided into those permissive also according to TCE4 (—), or those TCE4-nonpermissive in HvG ( ) or GvH (dash-dot line). Right panel: All HLA-DPB1 mismatched transplantations (n = 537), stratified according to the presence of TCE4-permissive (—) or TCE4-nonpermissive HvG (
) or GvH (dash-dot line). Right panel: All HLA-DPB1 mismatched transplantations (n = 537), stratified according to the presence of TCE4-permissive (—) or TCE4-nonpermissive HvG ( ) or GvH (dash-dot line) mismatches.
) or GvH (dash-dot line) mismatches.
The impact of nonpermissive HLA-DPB1 mismatches on OS was also analyzed in the subgroups matched for 10 of 10, 9 of 10, or less than or equal to 8 of 10 of the HLA-A, B, C, DRB1, and DQB1 alleles. Concordant with our previous findings,11 nonpermissive DPB1 disparities defined according to TCE3 were significantly predictive of survival in the 10 of 10 matched pairs, with 2-year survival probabilities of 53% (43%-63%) versus 40% (29%-51%; P = .03; Figure 3A left panel). In addition, in these pairs, however, further classification of the TCE3-permissive mismatches into TCE4-permissive or TCE4-nonpermissive, demonstrated that the latter had a significantly lower probability of survival compared with the former (65% [52%-78%] vs 39% [24%-54%]; P = .02; Figure 3A middle panel). Note that the survival probabilities associated with nonpermissive mismatches classified as such in TCE4 but not in TCE3 were very similar to those associated with nonpermissive TCE3 disparities (40% vs 39%). In line with this, in the 10 of 10 matched pairs overall, nonpermissive HLA-DPB1 disparities according to TCE4 were highly predictive of OS, with 2-year survival probabilities of 65% (53%-78%) versus 40% (31%-48%) in TCE4-permissive pairs (P = .003; Figure 3A right panel). Importantly, TCE3 was not predictive of OS in 9 of 10 matched pairs (48% [38%-58%] vs 39% [29%-49%] in the TCE3 permissive and nonpermissive pairs; P = .42; Figure 3B left panel). Again, pairs classified as TCE3-permissive could be further subdivided into TCE4-permissive or TCE4-nonpermissive, and the latter had a significantly lower probability of survival compared with the former (55% [40%-70%] vs 41% [27%-55%]; P = .016; Figure 3B middle panel). As a result, TCE4 was significantly predictive of OS also in 9 of 10 matched pairs, with 2-year survival probabilities of 55% [40%-70%] versus 39% [31%-48%]; P = .02) in TCE4-permissive versus nonpermissive pairs (Figure 3B right panel). In contrast, the effect of nonpermissive HLA-DPB1 mismatches was abrogated by the presence of 2 or more mismatches at other HLA loci, both for TCE3 and TCE4 (37% [31%-43%] vs 39% [26%-52%]; P = .72 for TCE3 and 40% [25%-55%] vs 37% [27%-47%]; P = .61 for TCE4; Figure 3C left and right panel). Importantly, the survival estimates in patients transplanted from 10 of 10 or 9 of 10 matched donors with TCE4-nonpermissive HLA-DPB1 mismatches were similar to that of patients transplanted from less than or equal to 8 of 10 matched donors overall (40% vs 38%; P = .64).
Predictive value of TCE3 and TCE4 for OS after unrelated HSCT, stratified according to matching status at other HLA loci. Shown are Kaplan-Meier estimates of survival after HLA-DPB1 allele-mismatched transplantations, matched for 10 of 10 (A; n = 201), 9 of 10 (B; n = 199), or less than or equal to 8 of 10 (C; n = 137) of the alleles at HLA loci A, B, C, DRB1, and DQB1. (Left panels) Transplantations were stratified according to the presence of TCE3-permissive (solid lines) or nonpermissive (dashed lines) HLA-DPB1 mismatches. (Middle panels) TCE3-permissive transplantations were further subdivided into those permissive also according to TCE4 (—) and those nonpermissive according to TCE4 ( ). (Right panels) Transplantations were stratified according to the presence of TCE4-permissive (—) or TCE4-nonpermissive (
). (Right panels) Transplantations were stratified according to the presence of TCE4-permissive (—) or TCE4-nonpermissive ( ) HLA-DPB1 mismatches.
) HLA-DPB1 mismatches.
Predictive value of TCE3 and TCE4 for OS after unrelated HSCT, stratified according to matching status at other HLA loci. Shown are Kaplan-Meier estimates of survival after HLA-DPB1 allele-mismatched transplantations, matched for 10 of 10 (A; n = 201), 9 of 10 (B; n = 199), or less than or equal to 8 of 10 (C; n = 137) of the alleles at HLA loci A, B, C, DRB1, and DQB1. (Left panels) Transplantations were stratified according to the presence of TCE3-permissive (solid lines) or nonpermissive (dashed lines) HLA-DPB1 mismatches. (Middle panels) TCE3-permissive transplantations were further subdivided into those permissive also according to TCE4 (—) and those nonpermissive according to TCE4 ( ). (Right panels) Transplantations were stratified according to the presence of TCE4-permissive (—) or TCE4-nonpermissive (
). (Right panels) Transplantations were stratified according to the presence of TCE4-permissive (—) or TCE4-nonpermissive ( ) HLA-DPB1 mismatches.
) HLA-DPB1 mismatches.
The survival advantage mediated by HLA-DPB1 TCE4-permissiveness in 10 of 10 or 9 of 10 matched transplantations was observed not only in early disease stage patients (85% vs 48%; P = .004) but also, although less markedly, in advanced disease stage patients (47% and 35%; P = .02; Figure 4).
Association of nonpermissive HLA-DPB1 disparities according to TCE4 with increased mortality is seen both in early and in advanced disease. Kaplan-Meier estimates of survival after 10 of 10 or 9 of 10 allele-matched unrelated HSCT for early stage acute leukemia (AL-CR1, n = 81; left panel) or advanced disease (ADV, n = 296; right panel). Transplantations were divided into TCE4-permissive (—) or TCE4-nonpermissive HLA-DPB1 disparities ( ).
).
Association of nonpermissive HLA-DPB1 disparities according to TCE4 with increased mortality is seen both in early and in advanced disease. Kaplan-Meier estimates of survival after 10 of 10 or 9 of 10 allele-matched unrelated HSCT for early stage acute leukemia (AL-CR1, n = 81; left panel) or advanced disease (ADV, n = 296; right panel). Transplantations were divided into TCE4-permissive (—) or TCE4-nonpermissive HLA-DPB1 disparities ( ).
).
Cox and logistic regression analysis of OM, NRM, graft failure, aGVHD, and RI.
The survival advantage mediated by the presence of permissive rather than nonpermissive HLA-DPB1 TCE4 mismatches was also observed in unadjusted as well as adjusted Cox regression models of OM. Nonpermissive HLA-DPB1 TCE4 disparity was found to be a significant risk factor for OM (hazard ratio [HR] = 1.50; CI, 1.13-2.01; P = .005), independently from other significant non-HLA variables, including donor and patient gender, patient age, conditioning regimen (MA or RIC), stem cell source, and disease group (CML-CP1, AL-CR1, or ADV). In line with previous reports,26,27 the presence or absence of additional mismatches at HLA-DRB3/4/5 (HR = 0.97; CI, 0.69-1.40; P = .88), DQA1 (HR = 1.03; CI, 0.58-1.84; P = .91), and DPA1 (HR = 1.20; CI, 0.88-1.63; P = .25) did not have a significant impact on OM, neither in patients overall, nor in the subgroups of patients scored as TCE4-permissive or nonpermissive (data not shown). Taking 10 of 10 allele-matched, HLA-DPB1 TCE4-permissive transplantations as reference, the adjusted hazards of OM were significantly increased by the presence of TCE4-nonpermissive disparities in 10 of 10 and 9 of 10 allele-matched transplantations (Table 2).
Cox regression models for OM, NRM, and relapse and logistic regression models for graft failure and aGVHD
| . | Univariate models . | Multivariate models* . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OM† . | NRM† . | Graft failure† . | aGvHD 2-4‡ . | aGvHD 3-4‡ . | Relapse‡ . | OM† . | NRM† . | Graft failure† . | aGvHD 2-4‡ . | aGvHD 3-4‡ . | Relapse‡ . | |
| 10 of 10 allele-matched; HLA-DPB1 permissive | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 10 of 10 allele-matched; HLA-DPB1 nonpermissive | 2.04 (1.26-3.30) | 3.33 (1.58-7.01) | 5.79 (0.74-45.4) | 1.05 (0.52-2.11) | 1.82 (0.49-6.74) | 1.22 (0.69-2.16) | 2.12 (1.23-3.64) | 3.41 (1.45-8.02) | NA§ | 1.29 (0.58-2.87) | 1.82 (0.49-6.73) | NA‖ |
| P | .003 | .002 | .09 | .88 | .37 | .40 | .006 | .005 | .52 | .37 | ||
| 9 of 10 allele-matched; HLA-DPB1 permissive | 1.32 (0.72-2.40) | 2.48 (1.06-5.80) | 2.21 (0.19-25.1) | 1.83 (0.81-4.12) | 5.02 (1.31-19.2) | 1.21 (0.61-2.40) | 1.38 (0.71-2.69) | 2.14 (0.81-5.66) | NA§ | 1.75 (0.69-4.45) | 5.01 (1.31-19.2) | NA‖ |
| P | .36 | .04 | .52 | .14 | .02 | .58 | .33 | .12 | .24 | .02 | ||
| 9 of 10 allele-matched; HLA-DPB1 nonpermissive | 2.29 (1.42-3.69) | 4.18 (2.01-8.72) | 6.84 (0.88-52.9) | 1.86 (0.95-3.65) | 4.17 (1.21-14.4) | 1.25 (0.70-2.22) | 2.21 (1.28-3.80) | 3.69 (1.58-8.61) | NA§ | 2.11 (0.97-4.56) | 4.15 (1.20-14.3) | NA‖ |
| P | .001 | < .001 | .06 | .07 | .02 | .44 | .004 | .002 | .06 | .02 | ||
| No more than 8 of 10 allele-matched; HLA-DPB1 irrespective | 2.30 (1.43-3.71) | 3.93 (1.88-8.23) | 5.99 (0.76-47.0) | 1.78 (0.90-3.53) | 2.41 (0.67-8.68) | 1.30 (0.74-2.31) | 2.04 (1.18-3.54) | 3.06 (1.30-7.19) | NA§ | 2.13 (0.97-4.65) | 2.40 (0.66-8.65) | NA‖ |
| P | .001 | < .001 | .09 | .09 | .18 | .36 | .01 | .01 | .06 | .18 | ||
| . | Univariate models . | Multivariate models* . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OM† . | NRM† . | Graft failure† . | aGvHD 2-4‡ . | aGvHD 3-4‡ . | Relapse‡ . | OM† . | NRM† . | Graft failure† . | aGvHD 2-4‡ . | aGvHD 3-4‡ . | Relapse‡ . | |
| 10 of 10 allele-matched; HLA-DPB1 permissive | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 10 of 10 allele-matched; HLA-DPB1 nonpermissive | 2.04 (1.26-3.30) | 3.33 (1.58-7.01) | 5.79 (0.74-45.4) | 1.05 (0.52-2.11) | 1.82 (0.49-6.74) | 1.22 (0.69-2.16) | 2.12 (1.23-3.64) | 3.41 (1.45-8.02) | NA§ | 1.29 (0.58-2.87) | 1.82 (0.49-6.73) | NA‖ |
| P | .003 | .002 | .09 | .88 | .37 | .40 | .006 | .005 | .52 | .37 | ||
| 9 of 10 allele-matched; HLA-DPB1 permissive | 1.32 (0.72-2.40) | 2.48 (1.06-5.80) | 2.21 (0.19-25.1) | 1.83 (0.81-4.12) | 5.02 (1.31-19.2) | 1.21 (0.61-2.40) | 1.38 (0.71-2.69) | 2.14 (0.81-5.66) | NA§ | 1.75 (0.69-4.45) | 5.01 (1.31-19.2) | NA‖ |
| P | .36 | .04 | .52 | .14 | .02 | .58 | .33 | .12 | .24 | .02 | ||
| 9 of 10 allele-matched; HLA-DPB1 nonpermissive | 2.29 (1.42-3.69) | 4.18 (2.01-8.72) | 6.84 (0.88-52.9) | 1.86 (0.95-3.65) | 4.17 (1.21-14.4) | 1.25 (0.70-2.22) | 2.21 (1.28-3.80) | 3.69 (1.58-8.61) | NA§ | 2.11 (0.97-4.56) | 4.15 (1.20-14.3) | NA‖ |
| P | .001 | < .001 | .06 | .07 | .02 | .44 | .004 | .002 | .06 | .02 | ||
| No more than 8 of 10 allele-matched; HLA-DPB1 irrespective | 2.30 (1.43-3.71) | 3.93 (1.88-8.23) | 5.99 (0.76-47.0) | 1.78 (0.90-3.53) | 2.41 (0.67-8.68) | 1.30 (0.74-2.31) | 2.04 (1.18-3.54) | 3.06 (1.30-7.19) | NA§ | 2.13 (0.97-4.65) | 2.40 (0.66-8.65) | NA‖ |
| P | .001 | < .001 | .09 | .09 | .18 | .36 | .01 | .01 | .06 | .18 | ||
NA indicates not applicable.
Multivariate models include sex, age, and CMV status of donor and patient, year of transplantation, use of ATG, disease group, stem cell source, conditioning regimen, and use of TBI.
TCE4-nonpermissive mismatches were considered in the combined group of GvH and HvG direction and confronted with permissive mismatches. Numbers were for 10 of 10 allele-matched pairs: n = 140 nonpermissive, n = 61 permissive; for 9 of 10 allele-matched pairs: n = 145 nonpermissive, n = 54 permissive; for ≤8 of 10 allele-matched pairs: n = 137.
TCE4-nonpermissive mismatches were considered only in the GvH direction and confronted with the combined group of TCE4-permissive mismatches and TCE4-nonpermissive mismatches in the HvG direction. Numbers were for 10 of 10 allele-matched pairs: n = 72 nonpermissive, n = 129 permissive; for 9 of 10 allele-matched pairs: n = 85 nonpermissive, n = 114 permissive; for ≤8 of 10 allele-matched pairs: n = 137.
Not applicable because the number of events in each subgroup was too limited to allow statistically meaningful analysis.
Not applicable because the P > .2 in univariate models.
The increase in mortality risk was the result of a significant increase in NRM in adjusted models associated with nonpermissive TCE4 disparities (HR = 1.74; CI, 1.19-2.53; P = .004, overall and Table 2). In the total cohort, there was also a significant increase in the adjusted hazards of aGVHD grade 3 to 4 in the TCE4-nonpermissive GvH compared with the other groups (HR = 1.89; CI, 1.12-3.21; P = .02), which was however dependent on allelic mismatches at other loci because it was not evident in the separate analysis of the 10 of 10 or 9 of 10 allele-matched subgroups (Table 2). In addition, the hazards of relapse were not significantly different between the TCE4-nonpermissive GvH compared with the combined TCE4-permissive and HvG groups (HR = 1.02; CI, 0.73-1.42; P = .92, overall and Table 2). These findings suggest that nonpermissive TCE4 disparities, different from allelic HLA-DPB1 mismatches, do not significantly increase the risk of GVHD and do not enhance graft versus leukemia (GVL) activity. There was a trend for increased incidence of graft failure in nonpermissively mismatched transplantations (HR = 3.07; CI, 0.86-10.97; P = .08 in multivariate analysis overall, and Table 2 for univariate analysis).
Discussion
Following current national and international guidelines, unrelated HSC donor-recipient searches are primarily based on 4-digit typing for HLA-A, B, C, and DRB1 alleles because matching for these alleles has been shown to significantly improve clinical outcome in terms of OS, NRM, and aGVHD (http://www.marrow.org, http://www.ibmdr.galliera.it).28-30 ) Donor-recipient pairs high resolution matched for these 4 loci (8 of 8), in most instances, are also matched for HLA-DQB1 (10 of 10), resulting from strong linkage disequilibrium between DRB1 and DQB1 alleles.8 In contrast, HLA-DPB1 displays weak linkage disequilibrium with the other class II loci8 ; therefore, only approximately 15% of 10 of 10 matched pairs are also 4-digit allele-matched for HLA-DPB1 (12 of 12).31 It has recently been shown that matching for HLA-DPB1 is a double-edged sword because it significantly reduces NRM and aGVHD, but on the other hand increases the hazards of disease relapse, ultimately resulting in no significant advantage in OS.24,25 Our previous data have shown that in 10 of 10 matched pairs, group-specific rather than allele-specific HLA-DPB1 matching on the basis of T-cell alloreactivity patterns defining the 3-group algorithm TCE3, is significantly predictive of survival.11 This finding was confirmed in the present study. However, we also demonstrate that the TCE3 misclassifies approximately 50% of the permissive pairs involving HLA-DPB1*02 because these have a probability of survival as low as those classified as TCE3-nonpermissive (Figures 2, 3). As a result, the modified 4-group algorithm TCE4 we developed in the present study is markedly more predictive of OS than TCE3, both in 10 of 10 and in 9 of 10 matched pairs (Figure 3) and in the total cohort of all 537 informative patients (Figure 2). Our data demonstrate that survival probabilities can be significantly increased by selecting donors with TCE4-permissive HLA-DPB1 disparities. Importantly, this advantage was observed not only in patients transplanted with acute leukemia in first complete remission, in whom the impact of donor-recipient HLA matching status is known to be most pronounced, but also in patients with advanced disease at transplantation (Figure 4). On the basis of these data, we suggest that HLA-DPB1 TCE4 disparity should be characterized up-front in unrelated donor searches, to prospectively direct selection of potentially 10 of 10 or 9 of 10 matched donors toward those presenting TCE4-permissive HLA-DPB1 mismatches.
Although the observation that survival probabilities after unrelated HSCT can be significantly improved by “intelligent” donor selection based on avoidance of TCE4-nonpermissive HLA-DPB1 disparity is good news for patients, the bad news is that approximately 70% of HLA-DPB1 allelic mismatches found in white or Japanese donor-recipient pairs are predicted to score as TCE4-nonpermissive. This is because HLA-DPB1*02, classified as immunogenic in TCE4 but not in TCE3, has an allelic frequency of 20% in whites and Japanese.20,32 Based on this, the overall predicted probability for a given donor-recipient pair to be TCE4-permissively HLA-DPB1 mismatched is 26%, regardless of matching status at the other HLA loci. However, extension of the search to include both the 10 of 10 and the 9 of 10 allele-matched pool, which in our cohort was present for 400 of 537 (74%) transplantations, raises the chances of identifying a TCE4-permissive donor. Indeed, in our retrospective study of randomly selected donors with regards to HLA-DPB1, 115 of 537 (21.4%) of transplantations were performed from 10 of 10 or 9 of 10 matched, HLA-DPB1 TCE4-permissive donors (Table 1). This number might increase if HLA-DPB1 TCE4 permissiveness were to be included prospectively into the algorithms for donor selection.
It is interesting to note that, different from allelic HLA-DPB1 mismatches, TCE4-nonpermissive DPB1 disparities did not further enhance GVHD or GVL in our patients (Table 2). This suggests that NRM and GVHD/GVL may be governed by distinct immunologic mechanisms, an observation that deserves further investigation because it might have potential impacts on how to exploit HLA-DPB1 disparity for the prevention of disease relapse after unrelated HSCT.
The molecular nature of the epitope target of preferential T-cell alloreactivity directed against HLA-DPB1 is as yet largely elusive. The patient from whom the HLA-DPB1-specific T cells used to originally define nonpermissive disparities were derived was not informative for HLA-DPB1*02 because DPB1*0201 was shared between the patient and her stem cell donor and T cells specific for the antigen encoded by this allele are likely to have been deleted from the patient's repertoire.23 Consequently, the shared epitope recognized by cross-reactive alloreactive T cells on HLA-DP antigens from group 1 and 2 alleles can be predicted to be structurally different from the epitope recognized by HLA-DPB1*02-specific T cells. Interestingly, group 1 and 2 alleles share most amino acid residues in regions A and F of the HLA-DP beta chain and are in linkage disequilibrium with DPA1*02.33 In contrast, HLA-DPB1*02 has markedly different amino acid sequences in regions A and F and is found in linkage disequilibrium with DPA1*01, further supporting the notion that the relevant T-cell epitope encoded by this allele is substantially different from the other. The role of the DPα chain, encoded in cis or in trans, in formation of the relevant epitopes remains to be investigated. The HLA-DPA1 matching status in this study was not significantly predictive of survival; however, DPA1 typing was available for only 51% of the pairs. Moreover, the role of defined DPA1-DPB1 combinations remains to be investigated in larger transplantation cohorts.
Our data demonstrate that the approach used here to define matching algorithms on the basis of T-cell alloreactivity is a powerful strategy for the characterization of nonpermissive mismatches in unrelated HSCT. This objective is a subject of rising interest in the field because it has become increasingly difficult to find allele-level matched UDs, a result of the impressive degree of polymorphism unraveled by increasingly sophisticated HLA-typing techniques. Recently, different authors have attempted to define nonpermissive disparities by structural comparison of amino acid sequences encoded by mismatched alleles.9,34 The results of such approaches are dependent on the availability of high-powered statistical analysis, given the extreme complexity of this type of comparison. In addition, structural approaches probably miss immunogenic epitopes dependent on conformational mismatches as well as peptide-dependent epitopes, which have been shown to be relevant for T-cell alloreactivity, and are therefore more problematic than similar strategies used to define permissive mismatches for humoral alloreactivity in solid organ transplantation.35 The use of alloreactive T-cell cross-reactivity patterns circumvents these problems and might in the future be applied also for defining nonpermissive mismatches at other HLA loci.
Taken together, our data demonstrate that group-specific HLA-DPB1 matching according to TCE4 is markedly more predictive for the clinical outcome of unrelated HSCT than the original TCE3, showing a significant association with NRM and OS in 10 of 10 and 9 of 10 matched transplantations. These findings call for revisiting current concepts of unrelated donor-recipient matching, suggesting that UD searches should be directed up-front toward identification of a 10 of 10 or 9 of 10 matched donor presenting TCE4-permissive HLA-DPB1 disparities. Using this strategy, patients both in early and in advanced stage disease could potentially be offered a significantly improved chance of survival after transplantation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the Gruppo Italiano Trapianto di Midollo Osseo, CSE e terapia cellulare for its participation in this study.
This work was supported by grants from the Associazione Ialiana per la Ricerca sul Cancro (grant AIRC 44/2007), the Cariplo Foundation (grant 2007.5486), and the Telethon Foundation (grant GGP08201).
Authorship
Contribution: R.C. designed the study, collected data, and performed statistical analyses; E.Z and J.M. developed the algorithm for nonpermissive HLA-DPB1 disparities; L.V. prepared the figures and critically reviewed the manuscript; R.O. and B.B. collected clinical data; T.L., R.F., G.B., and A. Bosi provided advice and participated in general discussion; S.P. and N.S. collected immunogenetics data; M.P.S. supervised statistical analyses; L.G. and V.M. performed HLA typing; F.C. and A. Bacigalupo counseled on study design and participated in critical discussion; and K.F. supervised the study and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare that a patent application describing the results of the manuscript has been filed on behalf of the San Raffaele Scientific Institute and that the patent application has not been published yet.
A complete membership list of Gruppo Italiano Trapianto di Midollo Osseo, CSE e Terapia Cellulare and Italian Bone Marrow Donor Registry participants appears in the supplemental Appendix (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Correspondence: Katharina Fleischhauer, Immunogenetics Laboratory, Unit of Immunohematology and Blood Transfusion, Division of Regenerative Medicine and Stem Cell Therapy, San Raffaele Scientific Institute, via Olgettina 60, 20132 Milano, Italy; e-mail: fleischhauer.katharina@hsr.it.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal