Abstract
Abstract 3985
Poster Board III-921
Venous thrombosis has genetic and acquired risk factors, and it has been proposed that several risk factors are needed for the occurrence of the disease. The identification of common gene variants associated with venous thrombosis may improve the ability to predict the risk and understanding of this disease. In a recent study, we aimed to identify genetic variants that are associated with deep vein thrombosis in individuals aged younger than 70 years (JAMA 2008; 299:1306-14). Of nearly 20000 single nucleotide polymorphisms (SNPs) that were genotyped, 7 SNPs were associated with deep vein thrombosis (range odds ratios, 1.1-1.3). However, studies of thousands of SNPs can lead to false-positive associations. Replication studies are therefore pivotal to account for false-positive associations.
To assess the risk of venous thrombosis of aforementioned SNPs in a large population based study.
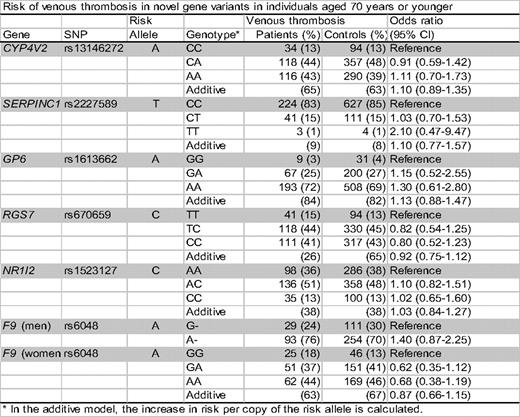
From the residents of Nord-Trøndelag county in Norway aged 20 years and older (n = 94194), we identified all patients with an objectively verified diagnosis of venous thrombosis that occurred between 1995 and 2001. By this date we had registered 515 patients with a first venous thrombosis; an age- and sex-stratified random sample of 1476 controls without previous venous thrombosis was drawn from the original cohort. Patients and diagnosis characteristics were retrieved from medical records. Of the 7 SNPs that were associated with deep vein thrombosis in our previous study, 6 were analyzed in the present study, i.e. rs13146272 in CYP4V2; rs2227589 in SERPINC1; rs1613662 in GP6; rs670659 in RGS7; rs1523127 in NR1I2; and rs6048 in F9. DNA concentrations were standardized to 10 ng/μL using PicoGreen (Molecular Probes, Invitrogen Corp, Carlsbad, CA, USA) fluorescent dye. Genotyping of individual DNA samples were demonstrated by polymerase chain reactions using the TaqMan assay. The technicians were blinded to whether the samples came from patients or control subjects.
The median age of both cases and controls at baseline was 70 years (range, 20-98). Almost half of patients and controls were men. Two thirds of the patients had deep vein thrombosis and one third had pulmonary embolism. Among the 515 events, 246 were idiopathic (48%). The prevalences of the 6 analyzed risk alleles were 17-96% in the patients and 16-96% in the control group. Only the F9 (rs6048) risk allele in men was consistently associated with an increased risk of venous thrombosis with odds ratios of 1.27 (95% CI, 0.83-1.93) for deep vein thrombosis, 1.62 (95% CI, 0.91-2.89) for pulmonary embolism, and 1.38 (95% CI, 0.97-1.97) for total venous thrombosis. For all other risk alleles we found odds ratios for venous thrombosis close to 1.0 compared to their reference allele. Odds ratios for provoked or idiopathic venous thrombosis again showed that only the F9 risk allele in men was consistently associated with an increased risk with an odds ratio of 1.30 (95% CI, 0.82-2.08) for provoked venous thrombosis and 1.47 (95% CI, 0.90-2.38) for idiopathic venous thrombosis, respectively, compared to men with the G allele. To make the current analysis more similar with our previous study, we restricted the analysis to those who were younger than 70 years. This resulted in slightly higher odds ratios for venous thrombosis in CYP4V2, SERPINC1 and GP6 variants (Table).
This population based study confirmed the previous finding that men with the A allele of rs6048 in F9 have an increased risk of venous thrombosis, while we could not replicate the association of gene variants in CYP4V2, SERPINC1, GP6, RGS7 and NR1I2 with venous thrombosis, possibly excepted for CYP4V2, SERPINC1 and GP6 gene variants in individuals aged younger than 70 years.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal