Abstract
Abstract 3300
Poster Board III-188
Imatinib mesylate (IM) has dramatically changed the management of chronic phase (CP) chronic myeloid leukemia (CML), with more than 80% of treated patients alive and free from transformation after 8 years of therapy. Long-term studies suggest that molecular response appears to increase in some patients over time and a significant minority of patients eventually obtain a “molecular remission”, where quantitative PCR (QPCR) for the bcr-abl transcript is negative. Two IM cessation trials have been conducted in a total of 90 patients who were consistently negative by QPCR. In each trial ∼40-50% of patients had molecular relapse after IM was stopped. Thus, despite the fact that IM, as well as dasatinib and nilotinib, do not appear to target CML stem cells, it appears that some patients may be able to stop IM therapy in the short-term, if not the long-term. These data suggest that different “kinetic” profiles exist: in some patients bcr-abl continues to decline, whereas in others it may plateau or even increase. In order to study this phenomenon, a more sensitive method of molecular detection is required. We utilized a nanofluidic platform (Fluidigm Corporation, South San Francisco, CA) to detect and quantify rare copies of bcr-abl mRNA. In this assay, detection sensitivity is augmented by sample partitioning prior to PCR, such that multiple QPCR reactions are performed simultaneously in hundreds of replicates. Initial bcr-abl copy number is calculated using the Poisson distribution. The assay was first optimized to reproducibly detect a single copy of bcr-abl using plasmids and bcr-abl positive cell lines in a background of bcr-abl negative cell lines. We then examined longitudinal samples (4-5 samples per patient) from 8 CP CML patients enrolled on the Novartis-sponsored TOPS trial who were in a molecular remission by QPCR. Among 33 patient samples, 7 samples were positive by QPCR and 26 samples were negative by QPCR. The Fluidigm nanofluidic QPCR assay was positive in all 7 samples positive by QPCR and was positive in 11 of 26 quantitative negative PCR samples (42%). Eleven of the 33 samples were also negative by a sensitive nested qualitative PCR assay performed in duplicate. Bcr-abl was detected using the nanofluidic QPCR assay in 1 of 11 of these samples (9%). Bcr-abl was not detected in 3 normal peripheral blood samples or in 3 bcr-abl negative cell lines. In one patient nanofluidic QPCR over time identified increasing bcr-abl 6 months before molecular relapse was evident by routine QPCR (Figure 1a,b ). In 5 patients bcr-abl decreased or was stable over time (Figure 1b ). In two patients persistently negative by both quantitative and nested qualitative PCR no bcr-abl was detected over time.
The nanofluidic QPCR assay can detect bcr-abl in ∼40% of cases negative by routine QPCR and can detect increasing bcr-abl copies many months before it is evident by routine QPCR testing. Interestingly, this detection frequency is similar to the relapse rate in molecular remission patients after IM is discontinued. This observation suggests the assay may be valuable in monitoring for molecular relapse, potentially in ongoing IM cessation trials.
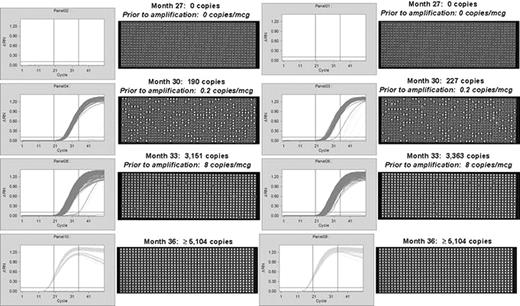
Quantitation of bcr-abl mRNA in quantitative negative PCR samples. For patient investigations up to 500 nanograms of total RNA was examined per panel. After a pre-amplification step consisting of reverse transcription followed by 13 cycles of PCR, each patient sample was loaded onto the nanofluidic chip. Each chip contains 12 sample inputs, each with 765 partitions. Bcr-abl was assessed in 2 to 8 panels (1,530 to 6,120 individual PCR reactions) for each patient. Negative and positive controls were present on each chip. RNA integrity was assessed using the control gene ABL. Copy numbers within the vertical lines (thresholds) are counted using the Poisson distribution (5,104 copies is the calculated maximum at saturation). Figure 1a. Data are shown for one patient in whom bcr-abl was detected using the nanofluidic QPCR assay 6 months prior to its detection by routine QPCR (see Figure 1b ). Because absolute quantitation occurs using the Fluidigm nanofluidic platform, copy numbers prior to amplification can be calculated per microgram (after adjustment for pre-amplification). Figure 1b. A comparison of bcr-abl kinetics in 2 patients by routine QPCR (bcr-abl copies) vs. nanofluidic QPCR (Poisson copies). For graphing purposes bcr-abl copy numbers are reported for both assays. All copy numbers > 5,100 are shown at 5,100.
Quantitation of bcr-abl mRNA in quantitative negative PCR samples. For patient investigations up to 500 nanograms of total RNA was examined per panel. After a pre-amplification step consisting of reverse transcription followed by 13 cycles of PCR, each patient sample was loaded onto the nanofluidic chip. Each chip contains 12 sample inputs, each with 765 partitions. Bcr-abl was assessed in 2 to 8 panels (1,530 to 6,120 individual PCR reactions) for each patient. Negative and positive controls were present on each chip. RNA integrity was assessed using the control gene ABL. Copy numbers within the vertical lines (thresholds) are counted using the Poisson distribution (5,104 copies is the calculated maximum at saturation). Figure 1a. Data are shown for one patient in whom bcr-abl was detected using the nanofluidic QPCR assay 6 months prior to its detection by routine QPCR (see Figure 1b ). Because absolute quantitation occurs using the Fluidigm nanofluidic platform, copy numbers prior to amplification can be calculated per microgram (after adjustment for pre-amplification). Figure 1b. A comparison of bcr-abl kinetics in 2 patients by routine QPCR (bcr-abl copies) vs. nanofluidic QPCR (Poisson copies). For graphing purposes bcr-abl copy numbers are reported for both assays. All copy numbers > 5,100 are shown at 5,100.
Radich:Novartis: Consultancy, Honoraria, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal