Abstract
Abstract 276
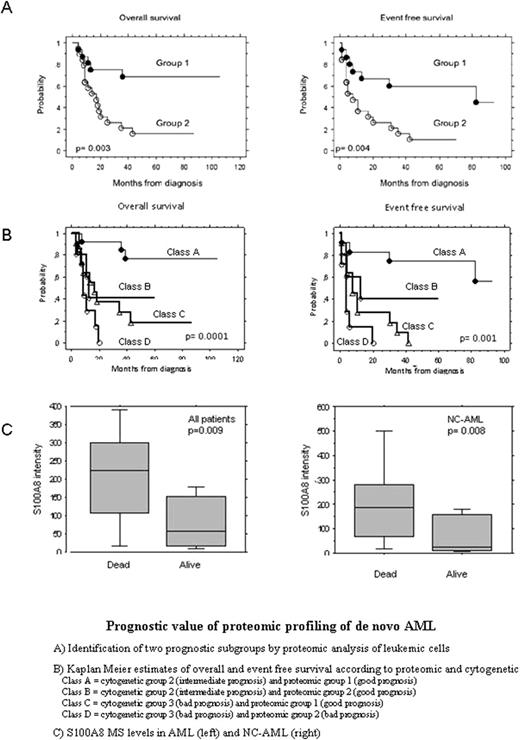
Treatment strategies of acute myeloid leukaemia (AML) largely depend on their cytogenetic status. However this stratification remains insufficient for almost half of the patients, requiring subsequent molecular investigations. In our study, we used mass spectrometry-based proteomic approaches to characterize de novo-AML. Samples (blood mononuclear cells collected and frozen before any treatment) were retrospectively selected from two independent sets of newly diagnosed AML patients, included in prospective clinical trials of the GOELAMS (Groupe d'Etude Ouest-Est des Leucémies aigues). We showed that protein signature of leukemic cells defined 2 groups of patients that displayed significant variation of overall and disease free survival (Fig-1A). This proteomic classification refined cytogenetic classes. Combination of proteomic and cytogenetic allowed a new stratification highly correlated with outcome. In particular, AML with intermediate and high risk cytogenetic could be respectively subdivided according to their protein profiles in two subgroups with significantly different survival (Fig-1B). Interestingly in both type 2 and type 3 cytogenetic groups, a good proteomic profile identified subsets of patients with significantly increased probability of survival, suggesting that the weigh of a good proteomic risk might predominate over a bad predictive cytogenetic. Among proteins expressed by leukemic cells, we isolated a 10800 Da marker that retained the highest discriminative value between alive and deceased patients. The median intensity of the 10800 peak was 94.3 ± 97.7 (7.5-368.7) in living patients compared to 214.2 ± 163.7 (9.0-693.4) in dead patients (p= 0.009). Among normal cytogenetic patients, intensity levels of the marker was also significantly different between dead (217.4 ± 155.7 range 10.4-612.8) and alive patients (26.0 ± 73.1, range 6-168.1; p= 0.008) (Fig-1C). Using a thresholds of 100 (calculated by ROC curves) we were able to correctly identify 82% of patients who died (patients greater than 100) and 70% of patients who stayed alive (patients lower than 100) with a specificity of 65% and 82% respectively. These data were confirmed in a second independent set of patients. 10800 Da marker was identified by MS peptide sequencing as S100A8 (also designated MRP8 or calgranulin A), a cytosolic protein of mature granulocytes. Western blot analysis confirmed its expression mainly in AML patients with the worst prognostic but not in all AML patients, neither in some leukemic and lymphoma cell lines, arguing for a selective deregulation associated with poor prognosis. These results show that expression of S100A8 in leukemic cells at diagnosis might be considered as a predictor of low survival.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal