Abstract
Abstract 2396
Poster Board II-373
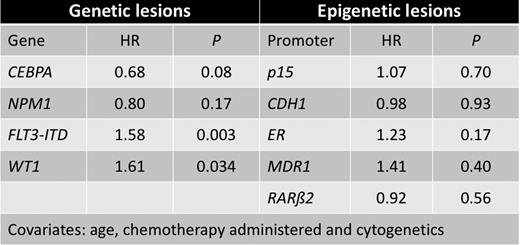
The prognosis of patients with acute myeloid leukemia (AML) depends largely upon the underlying cytogenetic- and molecular aberrations. We addressed the combined consequences of an epigenetic event, promoter hypermethylation, in five potential tumor suppressor genes (p15, CDH1, ER, MDR1 and RARb2) and mutational status in four genes with prognostic impact in adult AML (NPM1, CEBPA, FLT3 and WT1). Analyses were performed on pre-treatment samples from 70 pediatric and 378 adult AML patients. Children aged 0-15 years were enrolled from the two pediatric hematological departments in Denmark with tertiary function from 1987-2007, adults were treated at the Department of Hematology, Aarhus University Hospital, Denmark, between 1985 and 2007. Mutation status was determined either by fragment analysis followed by direct sequencing, or by sequencing alone. Methylation status was conducted with methylation-sensitive melting curve analysis (MS-MCA) after initial bisulphite modification. Comparisons of methylation or mutation frequencies between groups were done using chi-square tests. Wilcoxon's rank-sum test was employed to describe number of methylated genes in cytogenetically or mutationally defined subgroups. Survival analysis was done by Cox regression using the following covariates: age (either child, adult below 62 years or adult above 62 years), chemotherapy (either NOPHO regimen (Lie et al BJH 2003) (children), adult treated with curative intent (standard antracycline-cytarabine based), adult without curative intent (hydroxyurea or low-dose cytarabine) or no treatment) and cytogenetic status according to Grimwade et al (Blood 1998). To keep number of covariates at an acceptable level, mutation status and methylation status were analyzed separately. In pediatric AML cases, we found promoter hypermethylation in p15 (47%), CDH1 (64%), ER (62%), MDR1 (8%), and RARb2 (22%) and mutations in NPM1 (10%), CEBPA (3%), FLT3ITD (4%), and WT1 (7%). Comparing our results with data from the adult AML cohort we found significant differences between pediatric and adult patients for methylation in p15 (47 % vs. 73%, P<0.001) and RARb2 (22% vs. 42%, P=0.003), and for mutations in NPM1 (11% vs. 32%, P=0.001) and FLT3ITD (4% vs. 24%, P<0.001)), respectively. We found a strong association between promoter hypermethylation in p15, CDH1 and ER and core binding factor leukemias (CBF), and a negative correlation between methylation in the above genes, and abnormalities involving 11q23 (P=0.024) in pediatric AML. There was a trend towards this correlation in adult AML (P=0.094). Similarly, we found a higher number of methylated genes in patients lacking mutations in the NPM1, CEBPA, WT1 or FLT3 genes (P=0.026). We observed a trend for NPM1 and CEBPA mutations to be favorable prognostic factors (Hazard ratio (HR) 0.68 and 0.80, respectively, P=0.08 and P=0.17, respectively) while FLT3-ITD and WT1 mutation were found to be unfavorable prognostic factors (HR=1.58 and HR=1.61, P=0.003 and P=0.034, respectively). No survival effect was observed for the epigenetic aberrations probed for. (p15, HR=1.07, P=0.70, CDH1, HR=0.98, P=0.93, ER, HR=1.23, P=0.17, MDR1, HR=1.41, P=0.40 and RARb2, HR=0.92, P=0.56). In conclusion, in a large population-based AML cohort a direct comparison between childhood and adult AML revealed important biological differences. Furthermore, promoter hypermethylation occurs in a non-random pattern in AML depending on cytogenetic and mutational status, and might thus represent an important step in leukemogenic transformation, which is probably also dependent on patient age as epigenetic and genetic aberration frequencies vary in different age categories. These data confirm and extend recent studies in childhood AML and should constitute a platform for constructing improved prognostic systems in these patients.
Impact on survival of different genetic and epigenetic lesions. HR, Hazard ratio.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal